Using Bioinformatics to Study the Effects of COVID-19 on the Heart through Myocarditis
ABSTRACT
This research is addressing the problem with most COVID-19 vaccines. The problem with most COVID-19 vaccines is that after the patient gets the vaccine, their chances of getting a heart condition called myocarditis (the inflammation of the heart) is higher. The goal of this research is to understand the effects of COVID-19 on the heart through myocarditis as well as creating a vaccine which will not only be effective like the COVID-19 vaccine right now but also a much lower risk of myocarditis. There are many different ways to create vaccines, however, one of the most common ways is when the pathogen is isolated and grown until it loses its effectiveness in causing diseases. The weakest pathogens are then selected to put into the vaccine. Different bioinformatics tools and databases were used to collect, perform bioinformatics research experiments, analyze and interpret results. Using NCBI and GEO2R bioinformatics tool and database, we found 30 differentially expressed genes (DEGs) in the dataset GSE235433 with 15 genes being upregulated and 15 genes downregulated. The GO and KEGG pathway analysis showed that the upregulated DEGs were mainly involved in the signal transductor. From the DEGs we identified key genes with DAVID. The key DEGs were mainly related to 3 gene ontology terms.
INTRODUCTION
SARS-CoV-2 is a virus which causes the disease COVID-19. COVID-19 is very dangerous to the heart as it increases the chances of myocarditis substantially. Myocarditis is the inflammation of the heart and it can cause complications such as a heart attack, heart failure, and stroke (Myocarditis: Symptoms and Causes; Lasica et al, 2023). Myocarditis is a serious disease with high death and illness rates, especially in young people. A big challenge is that it is difficult to diagnose myocarditis accurately and quickly because there are many different causes and related diseases that can lead to it (Lasica et al, 2023).
Myocarditis is usually caused by an infection, but it can also result from drug hypersensitivity, radiation, metabolic disorders, collagen diseases, sarcoidosis, Kawasaki disease, and exposure to excessive heat or chemicals (Lasica et al, 2023).
It is difficult to know exactly how common myocarditis is because many people with the condition aren’t diagnosed. However, it’s estimated that between 10.2 and 105.6 out of every 100,000 people worldwide have it, with about 1.8 million new cases each year (Lasica et al, 2023).
During the COVID-19 pandemic, up to 28% of patients experienced heart damage, shown by increased levels of a heart-specific protein called troponin (Guo et al, 2020). A study found that 54% of patients who had COVID-19 showed signs of myocarditis (heart inflammation) on a cardiac MRI (Altay, 2022).
The scientific question being investigated in this research study is how SARS-CoV-2 is causing myocarditis in terms of gene expression and what can we do to prevent this from causing life threatening complications?
Almost everyone knows about COVID-19, however, most people only know about its effects on the lungs and the respiratory system (Galiatsatos et al., 2022), the effects COVID-19 has on the heart can be even more life threatening (Liu et al., 2020). The goal is to find how COVID-19 affects the heart and the effects of SARS-CoV-2 on the chances of myocarditis.
It is hypothesized that genes related to the heart will be differentially expressed in samples with the COVID-19 infection compared to samples without the infection. Therefore, the goals of this research are to first identify genes that are differentially expressed between samples that are COVID-19 infected and healthy samples. Then to identify genes related to the heart that resulting from SARS-CoV-2 that can be used to potentially create vaccines which do not cause an increase in the chances of myocarditis.
Throughout this study, many tools were used such as GEO (https://www.ncbi.nlm.nih.gov/gds/?term=), GEO2R, NCBI, and DAVID (https://david.ncifcrf.gov/tools.jsp). This research is important because using these DEGs and the analysis of them through Gene Ontology, we can create a vaccine to help protect people from COVID-19 as well as reduce the chances of myocarditis.
METHODS
Using NCBI to attain biological datasets:
The National Center for Biotechnology Information (NCBI) is the home to many biological datasets (https://www.ncbi.nlm.nih.gov/). NCBI allows us to analyze these datasets that we wouldn’t have been able to analyze before using its different features and tools (https://www.ncbi.nlm.nih.gov/geo/geo2r/).
In this study, NCBI was used and filtered out only GEO datasets so that when the dataset was found, the dataset would be able to be analyzed with GEO2R. Within GEO is the GEO2R that is a no-coding bioinformatics platform with different datasets of experiments based on RNA and genes (http://www.ncbi.nlm.nih.gov/geo/geo2r (Davis and Meltzer, 2007).
The GEO dataset we used was GSE235433. This dataset can be found on the NCBI website by searching up the exact dataset ID in the search bar. (GSE235433 )
Analyzing the Dataset with GEO2R and DAVID:
Once I found the dataset that pertained to my research topic, I then split the dataset into 9 different groups after which I clicked analyze with GEO2R. After analyzing the dataset with GEO2R, I was able to obtain differentially expressed genes (DEGs) and many graphs.
To select the top differentially expressed genes (DEGs),statistics were used via p value and log2 | fold change (FC) |. The P-value < 0.05 was considered to have statistical significance and to achieve significant differentially expressed genes. Log FC was used by selecting genes with a positive log FC and negative FC values (Supplementary Data: Gene IDs Differentially Expressed Genes ).
Identifying functions and biological pathways where genes are enriched using DAVID:
In order to predict and determine functions of genes and to identify where in the biological pathway certain genes are involved in, Gene Ontology (GO) functional terms (Ashburner et al, 2000) and KEGG databases are used (Kanehisa et al., 2012). Specifically, the GO terms for gene functions include biological process (BP), cellular component (CC), and molecular function (MF) ((Ashburner et al., 2000).
In this research study, I selected the top DEGs and analyzed them with the Database for Annotation, Visualization and Integrated Discovery (DAVID) bioinformatics database (https://david.ncifcrf.gov/) (Huang da et al., 2009) to perform the gene ontology and KEGG Pathway enrichment analysis (Kanehisa et al., 2012).
The main flow of steps in this research work is shown in the chart below. First, the research topic was found. Next, the geodataset related to the topic of research was found and then the data was analyzed with GEO2R. Then, based on the p-values specific DEGs were worked with and then DAVID was used to find the highly enriched terms through GO enrichment analysis.
Figure 1: Summary of steps used in this research work. Overview of the steps and methods used in this bioinformatics research work.
RESULTS
GEO2R Analysis:
When finding the DEGs that would like to be worked with, only the top DEGs that are statistically significant are selected. To do this, the top 15 upregulated (high enrichment) and top 15 down regulated (low enrichment) DEGs must be determined based on their p-values and log2 Fold change (Supplementary results: Gene IDs ). These gene IDs and p-values are obtained from the GEO2R analysis of the GEO dataset that is being worked with (GSE235433 ). After the GEO2R analysis is performed, graphs are created based on how the data is grouped.
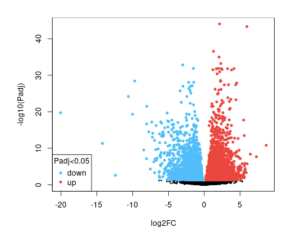
Figure 2a): The upregulated and downregulated genes in the infected timepoint 2 vs the infected timepoint 8 for the virus SARS-CoV-2. The blue dots represent the down regulated genes, while the red dots represent the up regulated genes. The black dots, however, represent genes that do not show any differences in expression between the control and infected samples.
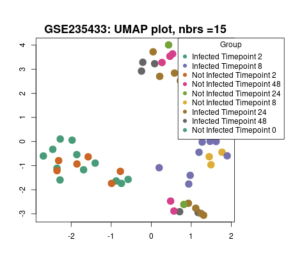
Figure 2b: UMAP Scatter plot containing genes of all 9 different groups. The closer each gene is to another, the more they are associated with each other.
Gene Ontology Enrichment Analysis:
The top 30 DEGs were found by selecting DEGs with less than 0.05 as the p-value and the top 15 DEGs with a positive LogFC value and top 15 DEGs with a negative LogFC value (Supplementary Results: Gene IDs ). Using these 30 top DEGs, the gene IDs of these DEGs can be pasted into DAVID (https://david.ncifcrf.gov/tools.jsp ) and then the gene ontology can be observed and it can be seen which terms are more enriched and which aren’t. Gene Ontology helps describe the biological domain as well as combine the gene and gene product attributes.
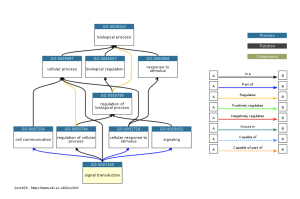
Figure 3a: The ancestry of the signal transduction GO term. The legend on the right of the chart shows what each arrow means and how each arrow relates to the flow of the ancestry chart. This chart shows the enrichment of the signal transduction term. The signal transduction is the pathway in which the COVID-19 virus travels through the cell (Rex et al., 2021).
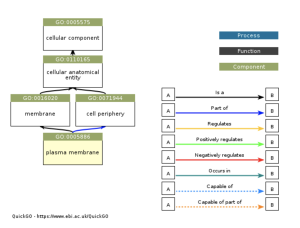
Figure 3b: The ancestry of the plasma membrane GO term. The legend on the right of the chart shows what each arrow represents and how each arrow relates to the origin of each part of the ancestry chart. This chart shows the enrichment of the plasma membrane term. The plasma membrane is involved with COVID-19 when COVID-19 stays along the plasma membrane the receptor molecules and fusion proteins bind together on the plasma membrane (Cohen et al., 2015).
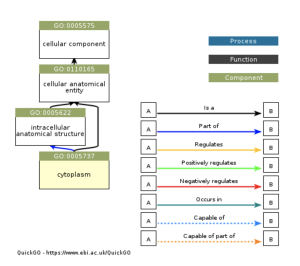
Figure 3c: The ancestry of the cytoplasm GO term. The legend on the right of the chart shows what each arrow represents and how each arrow relates to the flow of the ancestry chart. This chart shows the enrichment of the cytoplasm term. The cytoplasm is involved with COVID-19 when the COVID-19 cell replicates in the cytoplasm (Chen et al., 2022).
KEGG Pathway Enrichment Analysis:
The process for KEGG Pathway enrichment analysis is similar to the process for Gene Ontology enrichment analysis. The top DEGs are pasted into DAVID, and the KEGG Pathway can be observed and it can be seen which terms relate to the terms that are highly enriched in gene ontology and which terms don’t relate to them. The results showed that when analyzing any biological pathways with KEGG none of these DEGs showed significance involving the KEGG Pathway. There were no chart records of the KEGG Pathway involved with these DEGs.
Table 1) Summary of gene ontology terms that were highly enriched in the DEGs
Gene functions which were highly enriched | Specific gene ontology term |
Signal Transduction | Biological Process |
Plasma Membrane | Cellular Component |
Cytoplasm | Cellular Component |
The chart shown above shows the gene ontology terms that were highly enriched by performing the gene ontology enrichment analysis. Gene Ontology helps describe the biological domain as well as combine the gene and gene product attributes.
DISCUSSION
The main goal of this study was to identify and determine which differentially expressed genes in COVID-19 may be involved in the increased chances of myocarditis that the virus causes. To identify genes that are expressed, we used the Gene Expression Omnibus Bioinformatics database that is based on the R Programming language (https://www.ncbi.nlm.nih.gov/geo/). Then to identify the GO terms biological functions of the differentially expressed genes and to put these genes into corresponding biological pathways from the KEGG bioinformatics database, the Database for Annotation, Visualization and Integrated Discovery (DAVID) bioinformatics database (https://david.ncifcrf.gov/) was used (Huang da et al., 2009).
The results from performing the GEO2R analysis and selecting specific DEGs based on their p-values shows that these DEGs are highly associated with COVID-19 and its effects of causing an increase in chances of myocarditis. Using these DEGs, when we perform the gene ontology this process shows us that the terms signal transductor, plasma membrane, and cytoplasm are highly enriched which leads us to understand that COVID-19 associates with these terms the most (Figure 3). The gene ontology process also shows that of all the GO (gene ontology) terms, the cellular component is associated the most as it has more enriched terms and higher enriched terms (Table 1). This means that in terms of how these results relate to our research question, COVID-19’s association with the cellular component is one of the main causes of the increased chances of myocarditis.
This study’s results are similar to others such as this article from NCBI (Heidecker et al., 2022). As said in the article most patients have an increased chance of myocarditis after getting the vaccine, which is what we are trying to overcome as we are performing this research in an attempt to decrease those chances of myocarditis with a similarly effective vaccine.
The results produced by this research have great significance in developing a better vaccine for COVID-19. As of now, the COVID-19 virus has multiple vaccines, however, with most of these vaccines comes a higher chance of getting myocarditis (Heidecker et al., 2022). Using these results, however, the current COVID-19 vaccines have a possibility of working effectively along with the advantage of having a lower chance of getting myocarditis.
One limitation this research faced, however, was that all the genes that were used as a part of this research were part of bioinformatics datasets from other research, therefore, before creating a vaccine that reduces the chances of myocarditis, all the gene ontology terms identified must be studied in a laboratory or clinical setting.
The identified gene ontology terms can be tested in the laboratory by scientists in the laboratory or clinical trials to determine if vaccines can be made for COVID-19 without the chances of myocarditis being increased for future research.
Conclusion
In conclusion, throughout this study tools like NCBI (https://www.ncbi.nlm.nih.gov/) and DAVID (https://david.ncifcrf.gov/tools.jsp) were used to study gene expression in samples that are both infected and not infected by this virus. Through analysis with the GO bioinformatics database on the 30 DEGs, the highly enriched or associated gene ontology (GO) terms of the study were able to be understood. Results indicate that the signal transduction gene ontology term is the pathway in which the COVID-19 virus travels through the cell. Further, the results indicate that the plasma membrane is involved with COVID-19 when COVID-19 stays along the plasma membrane the receptor molecules and fusion proteins bind together on the plasma membrane. The cytoplasm is involved with COVID-19 when the COVID-19 cell replicates in the cytoplasm. The goal of this research was to understand the effects of this virus (COVID-19) on the heart through myocarditis and this study summarizes how COVID-19 potentially affects the heart through myocarditis. With these results from Table 1, researchers can potentially develop a vaccine that does not have the effects of an increased chance of myocarditis.
SUPPLEMENTARY DATA
References
- Altay S. COVID-19 myocarditis cardiac magnetic resonance findings in symptomatic patients. Acta Radiol. 2022;63:1475–1480. doi: 10.1177/02841851211046502.
- Chen, M., Ma, Y., & Chang, W. (2022). Sars-cov-2 and the nucleus. International Journal of Biological Sciences, 18(12), 4731–4743. https://doi.org/10.7150/ijbs.72482
- Cohen, F. S. (2016). How viruses invade cells. Biophysical Journal, 110(5), 1028–1032. https://doi.org/10.1016/j.bpj.2016.02.006
- Covid’s damage lingers in the heart | harvard medicine magazine. (n.d.). Retrieved June 20, 2024, from https://magazine.hms.harvard.edu/articles/covids-damage-lingers-heart
- David functional annotation bioinformatics microarray analysis. (n.d.). Retrieved June 20, 2024, from https://david.ncifcrf.gov/
- Guo T., Fan Y., Chen M., Wu X., Zhang L., He T., Wang H., Wan J., Wang X., Lu Z. Cardiovascular Implications of Fatal Outcomes of Patients with Coronavirus Disease 2019 (COVID-19) JAMA Cardiol. 2020;5:811–818. doi: 10.1001/jamacardio.2020.1017
- Geo2r—Geo—Ncbi. (n.d.). Retrieved June 20, 2024, from https://www.ncbi.nlm.nih.gov/geo/geo2r/?acc=GSE235433
- Heart problems after covid? Yes, it’s possible. (n.d.). Cleveland Clinic. Retrieved June 20, 2024, from https://my.clevelandclinic.org/health/articles/heart-problems-after-covid
- Heidecker, B., Dagan, N., Balicer, R., Eriksson, U., Rosano, G., Coats, A., Tschöpe, C., Kelle, S., Poland, G. A., Frustaci, A., Klingel, K., Martin, P., Hare, J. M., Cooper, L. T., Pantazis, A., Imazio, M., Prasad, S., & Lüscher, T. F. (2022). Myocarditis following COVID‐19 vaccine: Incidence, presentation, diagnosis, pathophysiology, therapy, and outcomes put into perspective. A clinical consensus document supported by the Heart Failure Association of the European Society of Cardiology (Esc) and the ESC Working Group on Myocardial and Pericardial Diseases. European Journal of Heart Failure, 10.1002/ejhf.2669. https://doi.org/10.1002/ejhf.2669
- Lasica R, Djukanovic L, Savic L, Krljanac G, Zdravkovic M, Ristic M, Lasica A, Asanin M, Ristic A. Update on Myocarditis: From Etiology and Clinical Picture to Modern Diagnostics and Methods of Treatment. Diagnostics (Basel). 2023 Sep 28;13(19):3073. doi: 10.3390/diagnostics13193073. PMID: 37835816; PMCID: PMC10572782.
- Liu, P. P., Blet, A., Smyth, D., & Li, H. (2020). The science underlying covid-19: Implications for the cardiovascular system. Circulation, 142(1), 68–78. https://doi.org/10.1161/CIRCULATIONAHA.120.047549
- Redirect notice. (n.d.-a). Retrieved June 20, 2024, from https://www.google.com/url?q=https://www.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2018.01093/full%23B45&sa=D&source=docs&ust=1718745276853694&usg=AOvVaw02bNVIGEQuaTYwpspkTaoW
- Redirect notice. (n.d.-b). Retrieved June 20, 2024, from https://www.google.com/url?q=https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8523273/&sa=D&source=docs&ust=1718745276853892&usg=AOvVaw3gvqQLyp3eL571TlAPO-DB
- Rex, D. A. B., Dagamajalu, S., Kandasamy, R. K., Raju, R., & Prasad, T. S. K. (2021). SARS-CoV-2 signaling pathway map: A functional landscape of molecular mechanisms in COVID-19. Journal of Cell Communication and Signaling, 15(4), 601–608. https://doi.org/10.1007/s12079-021-00632-4
- Sanbomics. (2022, January 8). Simple gene ontology and pathway enrichment from a gene list. https://www.youtube.com/watch?v=XLRA0A5qsoE
- Severe lung infection during COVID-19 can cause damage to the heart. (2024, March 20). National Institutes of Health (NIH). https://www.nih.gov/news-events/news-releases/severe-lung-infection-during-covid-19-can-cause-damage-heart
- Wang, S.-J., Brodie, K. C., De Pons, J. L., Demos, W. M., Gibson, A. C., Hayman, G. T., Hill, M. L., Kaldunski, M. L., Lamers, L., Laulederkind, S. J. F., Nalabolu, H. S., Thota, J., Thorat, K., Tutaj, M. A., Tutaj, M., Vedi, M., Zacher, S., Smith, J. R., Dwinell, M. R., & Kwitek, A. E. (2022). Ontological analysis of coronavirus associated human genes at the covid-19 disease portal. Genes, 13(12), 2304. https://doi.org/10.3390/genes13122304
- Zhuang, Z., Zhong, X., Chen, Q., Chen, H., & Liu, Z. (2022). Bioinformatics and system biology approach to reveal the interaction network and the therapeutic implications for non-small cell lung cancer patients with covid-19. Frontiers in Pharmacology, 13, 857730. https://doi.org/10.3389/fphar.2022.857730
- Vaccines. (2024, February 14). https://www.hopkinsmedicine.org/health/treatment-tests-and-therapies/vaccines



