Using Bioinformatics to Find Differentially Expressed Genes In Patients Using SGLT2i
_Using Bioinformatics to Find Differentially Expressed Genes In Patients Using SGLT2i - Diana Wang
ABSTRACT
Background
Heart disease or cardiovascular disease.is one of the most prevalent diseases in the United States. Almost half of all Americans have heart disease. A lot of lifestyle choices can affect someone’s risks for getting cardiovascular disease including family genetics. Recently, several genes that are linked to heart disease have been identified including new genes that increase the risk of heart disease.
Purpose
The problem our research is addressing is the lack of a cure for heart disease. Recent studies show that SGLT2i inhibitors, a medicine used to treat patients with diabetes, reduced people’s risk for getting heart disease.
Given the role of genes in inherited heart disease, the goal of this research is to identify additional genes, gene functions and biological pathways that can be potentially used to create new treatment solutions for heart disease such as gene therapy.
Methods
In our study, we started off by finding a dataset in GEO2R, a website that allows you to analyze data from various datasets. After separating the data into two groups; SGLT2i in use and SGLT2i not in use, we could further analyze the dataset. From there, we inserted the list of genes into a sheet and found the top 30 DEGs by sorting the list of genes into the genes with the highest log2FC value and lowest log2FC value.
The bioinformatics tool we chose to use to conduct analysis of our DEGs was SRplot. Then we inserted the gene symbols and log2FC values that corresponded to the symbols into SRplot.
After that, we decided to return back to the GEO2R dataset and included more patients into the two groups. By doing that, we could see which genes were important and compare the results. We found that there are three genes that are of importance in this study. Those three genes are also part of the top 30 DEGs.
Results
We chose to identify 30 DEGs. In total, 79 of the 17,925 genes were differentially expressed. There were 24 genes that were downregulated and 51 genes that were upregulated.
We found that histone lysine demethylation, histone demethylation, protein demethylation, protein dealkylation, histone demethylase activity, protein demethylase activity and demethylase activity have high enrichment scores.
We went back a second time to the GEO2R dataset and this time, included more patients into the two groups; stgl2i in use and stgl3i not in use so the results could be more accurate and it narrowed down the DEGs from 79 to 3. From there we could see that only 3 genes are differentially expressed. SCD, TSIX, and XIST which shows their significance. Then we went back to the google sheet, where we found that the SCD, TSIX, and XIST genes were all part of the top 30 DEGs. The three genes were all downregulated.
Discussion
This study shows that differentially expressed genes were identified between samples who received SGLT2i treatment and those patients who did not. The genes of significance were SCD, XIST and TSIX. The SCD gene, also known as Stearoyl-CoA Desaturase, helps produce fats that benefit the function of cell structures. XIST and TSIX genes work together to regulate XCI, an X chromosome in women.
My study provides a new direction to the development of a medicine or treatment because researchers can look into the genes of significance and SGLT2i’s impact on patients with Cardiovascular disease and try to create a treatment.
INTRODUCTION
Right now, heart disease is the leading cause of death (12). Smoking, drinking alcohol, and diets high in sugar are risk factors for heart disease. Poor diets can lead to heart disease.(9) Heart disease is a disease that affects the heart and its major blood vessels. Coronary artery disease is one of the most prevalent and common types of heart disease (7) . This happens when there is a lot of buildup in central blood vessels. This buildup prevents oxygen in the bloodstream from reaching the heart. When the blood vessel becomes nearly occupied with the buildup, the person is at risk for a heart attack or stroke. (20) Blockage in a vessel prevents oxygenated blood from reaching the heart.(1) . Although we still do not know the exact cause of heart disease (20) we can identify some risk factors which increase the risk of having heart disease (20) . Poor diets, lack of exercise, family genetics, and alcohol can affect someone’s risk for heart disease. How heart disease affects women has not been studied in depth yet. (22) Research has shown that hormones can affect a woman’s risk for heart disease.(14) Since heart disease is different in women, than it is in men, it can lead to misdiagnosis and plenty of other issues. (14)
Our study focuses on family genetics as the cause of heart disease In regard to family. Recently, an international research team, including the CARDioGRAM and the Coronary Artery Disease Genetics Consortia, confirmed ten genetic markers already linked to heart disease (11). In addition, they discovered 13 new ones that increase the risk of heart disease (11). Genetics play a complex role in heart disease. (3) Research shows that the risk of having heart disease increases in people with more than one sibling with cardiovascular disease. (3)
Therefore, understanding how these and additional genes function is important in helping figure out how the disease develops and could potentially lead to finding new medications or treatments including gene therapy.
SGLT2i is a term used to describe a class of different medications used to treat type 2 diabetes (18). Also known as Sodium Glucose Co-Transporter 2, these treatments are used to lower blood sugar levels, and recent studies show that SGLT2 inhibitors, which treat patients with type 2 diabetes, can actually help with cardiovascular disease (6) Common SGLT2 inhibitors include canagliflozin and dapagliflozin (18).
Gene therapy is the modification of a gene to form a cure. (2) Because of the role that genes play in family heart disease and the potential for gene therapy as a treatment for heart disease, the goal of this research is to find and compare genes in the control group to genes in the group using the medicine to see the similarities and differences. We hypothesize that there will be differences in expression of genes between the control group and patients.
Our research is important because as of now, there is still no cure for heart disease.(20) Yet heart disease continues to be the #1 cause of death.(12) By looking at how Stgl2i, a group of treatments designed to reduce glucose levels in the bloodstream for patients with diabetes, affects someone’s risk of heart disease, we can learn a lot about heart disease prevention. (6) Identifying the differences in gene expression between patients who don’t take the treatment compared to those who take the medicine can teach us a lot about ways to reduce the risk of having cardiovascular disease.
Results from this research with gene expression may help guide other researchers and scientists on the search to find a cure for heart disease or a treatment to reduce the risk of having heart disease.
METHODS
This study used different bioinformatics tools and databases, namely, GEO2R (8) and SRPlot (1) as indicated in Figure 1. The dataset we used was GSE26364.
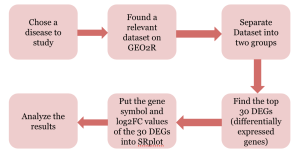
Figure 1. Flowchart showing methods and bioinformatics used in this study.
GEO and GEO2R Bioinformatics Analysis
We started off by using GEO, Gene Expression Omnibus (8), a bioinformatics tool that allows you to view other research experiments. We found an experiment similar to the topic we were studying, cardiovascular disease. From there we were able to go to GEO2R(8), and view the different patients. Then we proceeded to group the information into two groups; patients that did not use SGLT2i and patients that did use SGLT2i. After sorting the data into the two groups, we were able to analyze that data and view the different graphs. In this study, we tested to see if patients using SGLT2i had different expressed genes than patients not using SGLT2i. We tested 30 DEGs in total. 15 of the DEGs consisted of the highest log2FC value, while the rest were the lowest log2FC values. By going back to GEO2R (8) a second time to reanalyze the control and experimental group, we were able to narrow down the DEGs to find the genes that were the most significant in this study.
SRPlot Analysis of top DEGs
After finding my 30 DEGs, we were able to go to SRPlot (1), a website that allows you to enter the gene symbols of all the DEGs, and creates charts and graphs based off the symbols.
GO Analysis
Gene Ontology (10) is a bioinformatics website to assist in describing the function of genes and the gene functions. This tool allows us to describe genes in detail. The three main categories of the bioinformatics tool are; BP (biological processes), CC (cellular component) and MF (molecular function).
KEGG Analusis
KEGG (15), a group of databases that deal with genomes, biological pathways, diseases, and chemical substances. This tool is used for bioinformatics researchers to understand the biological functions of cells.
Top 30 DEGs – Top 30 DEGs
RESULTS
Analyzing Differential Gene Expression using GEO2R
The GEO2R (8) tool that generated our results used R programming through AL and ML algorithms. This R script explains the process and is lined here – R Script
We first went to GEO (8) and found a dataset that was relevant to this topic. We chose GSE26364. From there we were able to view the dataset and separate it into two groups; SGLT2i in use and SGLT2i not in use. We decided to use my top 30 DEGs to further analyze. From there we placed the gene symbols of the top 30 DEGs into SRPlot (1).
Results from GEO2R
In Figure 2, we identified 79 DEGs total out of 17,925 genes. A majority of the genes were upregulated.

Figure 2A. Venn Diagram In this figure, 79 of the 17,925 genes were differentially expressed.
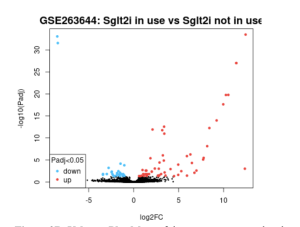
Figure 2B. Volcano Plot More of the genes are upregulated as shown on the graph. The blue dots represent downregulated genes and the black dots represent genes expressed the same in both groups
Results From GEO2R (second time)
Using GEO2R (8), the control and experimental groups were analyzed a second time and results showed, only 3 genes were found to be differentially expressed in Figure 3. The SCD, TSIX, and XIST genes were differentially expressed which shows their significance in this study. 3 of the 18121 genes were differentially expressed in Figure 4. Going back to the google sheet, we found that the SCD, TSIX, and XIST genes were all part of the top 30 DEGs.
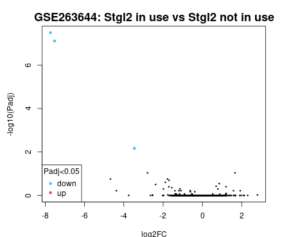
Figure 3. Volcano Plot Only 3 genes are downregulated.

Figure 4. Venn Diagram 3 of the 18,121 genes are differentially expressed.
Results from SRplot
The bioinformatics tool we chose to use to conduct analysis of our DEGs was SRplot (1). We inserted gene symbols and log2FC values of the top 30 DEGs into SRplot. Histone lysine demethylation, histone demethylation protein, demethylation histone, demethylase activity, and protein demethylase activity all had higher enrichment scores based on Figure 5. Overall, the enrichment scores were greater than or around 2. The CC section had the lowest enrichment scores compared to the BP and MF in Figure 5. In total, we found three significant genes. SCD, XIST and TSIX (Figure 6).
Results From SrPlot
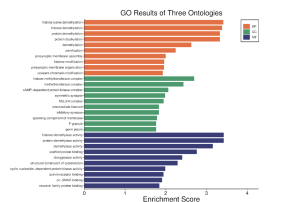
Figure 5. GO Results for BP, CC, MF Histone lysine demethylation, histone demethylation, protein demethylation, protein dealkylation, histone demethylase activity, protein demethylase activity and demethylase activity all have the higher enrichment score.
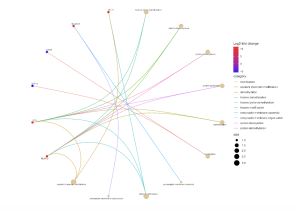
Figure 6. The size of the circles represents how significant they are. The lines connect the dots to show the relationship between genes and their biological function.
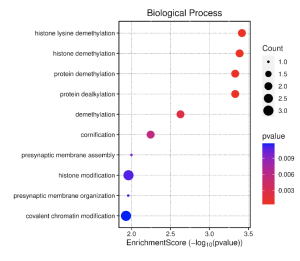
Figure 7. Dot Plot The greater the red of the circles means that the process has a lower p-value. The greater the size of the dots, the greater the significance.
Table 1. Summary of significant genes found in this study and their connection to heart disease
Gene | Function | Name | Connections to Heart Disease |
|---|---|---|---|
SCD | -Maintains homeostasis. -Keeps balance of the fats in a cell. | stearoyl-CoA desaturase | If SCD activity is reduced, the risk for cardiovascular disease increases and cardiovascular health is greatly affected. |
XIST | -Work with TSIX to regulate one X chromosome | X inactive specific transcript | Disruptions with the expression of XIST have been correlated/linked to cardiovascular disease. |
TSIX | -Work with XIST to regulate one X chromosome | TSIX X Inactive Specific Transcript | Problems with the regulation of the X chromosome could lead to issues with gene expression in cardiovascular processes. |
Supplementary Materials
R Script: R Script
Top DEGs: Top 30 DEGs
DISCUSSION
In this research, our goal was to identify genes, gene functions and biological pathways that can be potentially used to create new treatment solutions for heart disease such as gene therapy. We hypothesized that there would be differences in gene expression between patients using SGLT2i and patients not using SGLT2i.
This study is important because it could be a potential step towards finding a solution to the disease. We learned that although patients using the SGLT2i treatment could have differentially expressed genes (Figures 2, 3, 4, 5) , the treatment didn’t have a direct impact on the DEGs. We found DEGs between samples who received SGLT2i treatment and those who did not. We chose to look at 30 DEGs. In total, we found three significant genes. SCD, XIST and TSIX (Figure 6). Histone lysine demethylation, histone demethylation, protein demethylation, and protein dealkylation were pathways that had high enrichment scores (Figure 5). I found that all the genes of significance have a connection to heart disease.
Our findings in Figure 5. Figure 7 and Table 1 were similar to other studies in this field. In one study, STGL2i proved to positively benefit cardiovascular levels by preventing DNA demethylation in promoter regions. (19) In Figure 5, we can see that histone demethylation and protein demethylation had higher enrichment scores. In Figure 7, we can see that histone lysine demethylation, histone demethylation, protein demethylation are of greater significance and all have low p-values. In our study, we found a number of DEGs between samples who received Stgl2i treatment and others that did not (Figure 2 and Figure 4). The results show that while SGLT2 inhibitors have a positive effect on cardiovascular risk, the treatment was not directly linked to the difference in gene expression.
In another study, SGLT2 inhibitors had positive effects on the functions of the heart like decreased oxidative stress, improved energy metabolism, decreased epicardial fat mass, etc. (15) All these benefits eventually led to ‘decreased severity of heart failure. (15) While the diabetes treatment had various benefits on the body and heart, the treatment has no direct links to differences in gene expression.
In another research article, gene therapy could be a step closer to learning about a treatment for cardiovascular disease. (2) Since we identified DEGs between the two groups, other researchers could look into gene therapy as a treatment option for cardiovascular disease. Looking into how SGLT2 inhibitors lowers cardiovascular risk could also help with the search for finding a potential cure in the future.
In this case, the study was limited because all the information gathered was from a dataset on GEO2R (8). This dataset comes from an experiment done by other scientists. Further research will need to be completed on these identified genes in a laboratory before a potential treatment or cure may be found in the future.
By looking at all our results, we can conclude that there will be differences in gene expression between patients using SGLT2i and patients not using SGLT2i by viewing Figure 2 and 5. In Figure 2, we can see a few genes that are skewed to the right of the diagram. This skew indicated a difference in gene expression between the two groups. In Figure 5, a couple biological processes had higher enrichment scores. Those biological processes were histone lysine demethylation, histone demethylation, protein demethylation, and protein dealkylation, meaning they were significantly represented in the study. All these processes were of significance to our study. By identifying these important processes between the two groups, it indicated that there must have been differences in gene expression between patients using SGLT2i and patients not using SGLT2i.
Conclusion
This study provides new potential directions for the development of a potential cure because this study targets for the research of heart disease, which currently is one of the most prevalent diseases in the United States. By finding genes of significance, other researchers can further look into these genes and search for a correlation between genes and cardiovascular disease.
In this study, DEGs were identified between samples who received SGLT2i treatment and those who did not. Genes like SCD, XIST, and TSIX were significant in our study and XIST was found to have a direct link to cardiovascular disease. The top DEGs were enriched in biological pathways and molecular functions. We were able to identify genes with significant relationships with biological processes. The genes and biological processes can be studied in the lab for potential cure in the future.
References
- Bioinformatics (n.d.). Retrieved August 6, 2024, from https://www.bioinformatics.com.cn/en/
- Cannatà, A., Ali, H., Sinagra, G., & Giacca, M. (2020a, May 7). Gene therapy for the Heart Lessons Learned and future perspectives | circulation research. AHA ASA Journals. https://www.ahajournals.org/doi/10.1161/CIRCRESAHA.120.315855
- Czepluch, F. S., Wollnik, B., & Hasenfuß, G. (2018, June). Genetic determinants of heart failure: Facts and numbers. ESC heart failure. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5933969/
- Dahlöf, Björn. “Cardiovascular disease risk factors: epidemiology and risk assessment.” The American journal of cardiology 105.1 (2010): 3A-9A. www.sciencedirect.com/science/article/abs/pii/S0002914909024825.
- D’Agostino Sr, Ralph B., et al. “Cardiovascular disease risk assessment: insights from Framingham.” Global heart 8.1 (2013): 11-23. www.sciencedirect.com/science/article/pii/S2211816013000057.
- FayeRiley. “Type 2 Diabetes Medications Can Also Protect Heart Health.” Diabetes UK, Diabetes UK, 16 Feb. 2022, www.diabetes.org.uk/about-us/news-and-views/type-2-diabetes-medications-can-also-protect-heart-health.
- Franczyk B, Frąk W, Lisińska W, Młynarska E, Rysz J, Wojtasińska A. “Pathophysiology of Cardiovascular Diseases: New Insights into Molecular Mechanisms of Atherosclerosis, Arterial Hypertension, and Coronary Artery Disease.” Biomedicines. 2022 Aug 10;10(8):1938. doi: 10.3390/biomedicines10081938. PMID: 36009488; PMCID: PMC9405799. www.ncbi.nlm.nih.gov/pmc/articles/PMC9405799.
- “GEO2R: Analyze GEO Datasets.” National Center for Biotechnology Information, National Center for Biotechnology Information, https://www.ncbi.nlm.nih.gov/geo/info/geo2r.html. Accessed 1 Aug. 2024.
- Gatorworksdev. “How Does Diet Affect Your Heart Disease Risk?” Cardiovascular Institute of the South, 18 June 2024, www.cardio.com/blog/is-your-diet-increasing-your-heart-disease-risk.
- Gene Ontology Consortium. (2000). Gene Ontology: Tool for the unification of biology. Nature Genetics, https://geneontology.org/
- Hajar R. “Genetics in Cardiovascular Disease.” Heart Views. 2020 Jan-Mar;21(1):55-56. doi: 10.4103/HEARTVIEWS.HEARTVIEWS_140_19. Epub 2020 Jan 23. PMID: 32082505; PMCID: PMC7006335.
- “Heart Disease Facts.” Centers for Disease Control and Prevention, Centers for Disease Control and Prevention, 15 May 2024, www.cdc.gov/heart-disease/data-research/facts-stats/index.html.
- Heart Foundation. (n.d.). Family history and heart disease. Retrieved August 6, 2024, from https://www.heartfoundation.org.au/your-heart/family-history-and-heart-disease#
- Jhalani, Nisha. “Heart Disease in Women Is Not like Heart Disease in Men.” ColumbiaDoctors, 28 July 2023, www.columbiadoctors.org/news/heart-disease-women-not-heart-disease-men.
- Kyoto Encyclopedia of Genes and Genomes (1995) KEGG Pathway Database www.genome.jp/kegg.
- Lopaschuk, G. D., & Verma, S. (2020, June 22). Mechanisms of cardiovascular benefits of sodium glucose co-transporter 2 (SGLT2) inhibitors: A state-of-the-art review. JACC. Basic to translational science. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7315190/
- National Center for Biotechnology Information (NCBI). Programs & Activities. National Library of Medicine, 2023. www.ncbi.nlm.nih.gov/home/about/programs/.
- National Kidney Foundation. (n.d.). SGLT2 inhibitors. Retrieved August 1, 2024, from https://www.kidney.org/atoz/content/sglt2-inhibitors
- NCBI GEO. (n.d.). GEO2R: Analyze GEO Datasets. National Center for Biotechnology Information. www.ncbi.nlm.nih.gov/geo/info/geo2r.html.
- NHS. (n.d.). Cardiovascular disease. National Health Service. Retrieved August 1, 2024, from https://www.nhs.uk/conditions/cardiovascular-disease/
- Scisciola, L. (n.d.). Targeting high glucose-induced epigenetic modifications at cardiac level: The role of SGLT2 and SGLT2 inhibitors. Cardiovascular diabetology. https://pubmed.ncbi.nlm.nih.gov/36732760/
- “Women and Heart Disease.” National Heart Lung and Blood Institute, U.S. Department of Health and Human Services, www.nhlbi.nih.gov/health/coronary-heart-disease/women. Accessed 25 July 2024.
- Verhaar, M. C., E. Stroes, and T. J. Rabelink. “Folates and cardiovascular disease.” Arteriosclerosis, thrombosis, and vascular biology 22.1 (2002): 6-13. www.ahajournals.org/doi/full/10.1161/hq0102.102190.



