Using Bioinformatics in Order to Analyze the Top Differently Expressed Genomes with PKP2 Knockout from iPSC-CMs
ABSTRACT
Introduction
Arrhythmogenic right ventricular cardiomyopathy (ARVC) is a genetic heart disease that can lead to a higher risk of sudden death. Some symptoms are: irregular heartbeat, chest pain, and shortness of breath. The most common cause of ARVC is mutations, or changes in DNA and genes related to desmosomal proteins. These are structures in heart cells that help them stick together and function properly. Currently, there are no treatments available for this disease, and this research aims to help in the search for treatment. Many details about ARVC remain unclear which could possibly aid in the development of future treatments, such as new forms of gene therapy.
Methods
To conduct this research, five bioinformatics tools were used: NCBI, KEGG, GO (Gene Ontology), GEO2R, and DAVID (Database for Annotation, Visualization, and Integrated Discovery). DAVID was used to relate the genes in ARVC to other human diseases or potentially fatal causes of ARVC. Moreover, GEO2R was used to collect and analyze data from the data set GSE253223, which was a research study that tested two potential ARVC treatments with mouse models.The two treatments that categorized two groups referenced in the data set were siNEG and siPKP2. Finally, analysis by GEO2R helped identify differentially expressed genes (DEGs) in various samples sorted according to ARVC treatments, and DAVID was used to analyze GO and KEGG enrichment pathways to determine functions and biological pathways that the genes may be related to.
Results
Results from the GEO2R analysis identified over 18,000 genes, of which 252 were differentially expressed genes (DEGs) of the two treatment groups. These results were utilized by R programming, which was made by machine learning and artificial intelligence algorithms. It was unclear which genes were upregulated and downregulated, which suggests the need for further research. The DEGs were then narrowed down to the top 30 using the p-value in statistics. From the 252 DEGs, KEGG pathway analysis of the top 30 DEGs analyzed using DAVID showed that these DEGs were mostly associated with biological pathways: chemical carcinogenesis – reactive oxygen species, calcium signaling pathways, and TNF signaling pathways. Furthermore, the associated or enriched gene functions from the GO terms found phosphorylation to be the most enriched function, while ATP binding and calcium ion binding play indirect but important roles in ARVC. Three important genes of GSE253223 are DYNC1LI2, DDX10, and PDCD6, which were taken by the analysis of this dataset.
Conclusion
Results from this study can contribute to the broader field of personalized medicine or gene therapy where treatments and preventive strategies are tailored to an individual’s genetic makeup. The KEGG pathways, chemical carcinogenesis – reactive oxygen species, calcium signaling pathways, and TNF signaling pathways show how the genes included can relate to the cause of ARVC. The GO terms, phosphorylation, ATP binding, and calcium ion binding, can also contribute to the cause of ARVC.
KEYWORDS: Arrhythmogenic right ventricular cardiomyopathy (ARVC), Bioinformatics, NCBI GEO, DAVID, GO, KEGG
INTRODUCTION
Arrhythmogenic right ventricular cardiomyopathy, also known as ARVC, is a genetic heart disease that can cause irregular heartbeats and an increased risk of sudden death (Wu, Iris, et al. 2024). Currently, there is not much known about ARVC as it is a newer disease (Joshi-Mukherjee, Rosy, et al.2008). However, there is an understanding of its side effects, how it is genetic, and how it is mostly caused by mutations in the PKP2 gene, also known as plakophilin-2 (Stanford Health Care, 2017; Wu, Iris, et al. 2024). Around 40% of ARVC cases are related to changes or mutations in PKP2 (Wu, Iris, et al. 2024). PKP2 is responsible for creating a protein called plakophilin 2. Plakophilin 2 makes structures that are called desmosomes, which are formations in heart cells that help them stick together and function properly. When there is a lack of PKP2, ARVC will occur. Although there are many details about ARVC that are unclear, new bioinformatics research has added to our understanding by discovering a large number of implicated genes and pathways.
There are no known approved treatments that are able to address or heal the genetic causes of this disease (Chantal J.M., et al, 2019). Bioinformatics research can help find a potential cure or gene therapy for this disease. What is known so far is that ARVC is primarily caused by changes in DNA or genes called genetic mutations (Wu, Iris, et al. 2024; Chantal J.M., et al, 2019). These mutations often affect genes that code for proteins in the desmosomes. When these proteins are defective due to genetic mutations, heart muscle cells can weaken and deteriorate, particularly in the right ventricles, leading to irregular heartbeats and an increased risk of sudden death (Chantal J.M., et al, 2019; Wu, Iris et al, 2024).
Scientists hope that personalized medicine or gene therapy can someday be used to help address PKP2 gene mutations. To treat ARVC, scientists hope to use AAV9, also known as adeno associated virus 9, to deliver the normal non-mutated PKP2 gene that could possibly restore the PKP2 expression to help heal the heart (Wu, Iris, et al. 2024).
Therefore, the goal of this study is to identify additional genes, gene functions and biological pathways that can be used to create gene therapy solutions. The scientific question being investigated is: what genes are expressed differently when PKP2 is removed from heart cells, and what are the associated functions and biological pathways of these genes? The hypothesis is that there will be differences in gene expression between controls and patient samples since PKP2 is essential to making PKP2 protein to avoid ARVC.
This research study is important as it contributes to potential identifications of additional genes and biological pathways that can potentially be used to find a cure for ARVC involving gene therapy.
METHODS
Choosing a Human Disease
To conduct this research, several bioinformatics tools were used, namely, NCBI, GEO2R, DAVID, KEGG, and GO (Gene Ontology) (Figure 1). ARVC was chosen as the human disease to study because of the important need to understand the genetic causes of ARVC, develop treatments, and advance the public health knowledge of ARVC.
Finding a GEO Dataset on NCBI
The dataset that was chosen is under the accession number GSE253223, and it focuses on the effects of changing PKP2 levels on induced pluripotent stem cell derived cardiomyocytes (iPSC-CMs, also known as heart cells) over time. This dataset consists of 20 samples of homo sapiens, 4 of which have a treatment named siNeg, and the last 16 have a treatment called siPKP2 (plakophilin-2). All samples have the same cell type.
Finding the Top DEG’s With the P Value
To identify genes that are expressed differently in various samples, the samples were sorted into two groups. Then, the grouped data was analyzed using GEO2R, a bioinformatics tool, which resulted in over 50,000 identified differentially expressed genes (DEGs). Only the top 30 were statistically selected using the p-value. The DEGs were chosen with p-values < 0.05.
Analyzing the top DEG’s with KEGG and GO charts
To further analyze the selected top 30 DEGs to determine their functions and associated biological pathways, DAVID, a functional annotation tool, was used (david). This categorized the chosen genes into KEGG pathways and GO terms. KEGG pathway analysis identified important biological pathways, whereas GO enrichment analysis provided information about the biological processes associated with the DEGs. Finally, DAVID put the top 30 genes into KEGG (KEGG) and GO (2024-01-17).
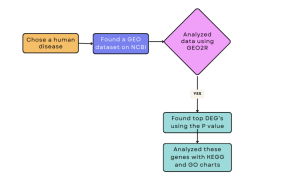
Figure 1: Methods Flowchart: Showing a summary of the methods used in this study to investigate PKP2 knockout in a mouse model.
RESULTS
Identification of Differentially Expressed Genes from GEO2R Analysis and Statistics
The DEGs that were obtained from GEO2R show how ARVC varies with each subject. Using the dataset, the top 30 DEGs were identified using the p-value (ARVC Top DEGs). All of the DEGs were upregulated from the data. GEO2R used R programming to analyze the results, which is pre-programmed with AI and ML (machine learning) algorithms. The R script detailing this can be found in this link.
Identification of Biological Pathways from DAVID Analysis
The biological pathways from the DAVID analysis show how some genes can be related to ARVC or other human diseases. Some of the pathways that were picked were the calcium signaling pathway, chemical carcinogenesis – reactive oxygen species, and the TNF signaling pathway. These three pathways were chosen for this research because they are the most related to ARVC.
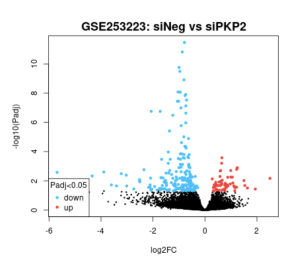
Figure 2: The volcano chart showing the upregulated and downregulated DEGs; the blue dots representing downregulation, and red dots representing upregulation. Black is neutral.
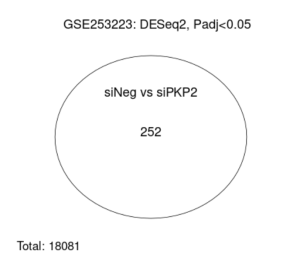
Figure 3: The venn diagram shows the similarities between the two treatments, siNEG and siPKP2
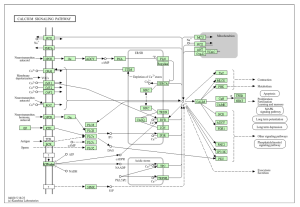
Figure 4: Shows calcium signaling In heart cells, necessary in order to maintain heart rhythm.
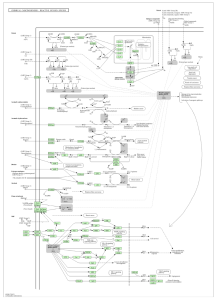
Figure 5: Chemical carcinogenesis involves oxidative stress leading to cancer. Even though PKP2 is not directly related, oxidative stress can impact cardiac cells and adhesion properties.
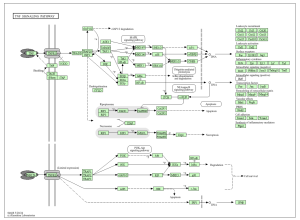
Figure 6: TNF signaling is involved with inflammation, cell survival, and immune responses. A certain cytokine in this pathway may promote inflammation in cardiac tissue, as well as fibrosis. This can lead to the replacement of normal heart muscle that contributes to heart failure.
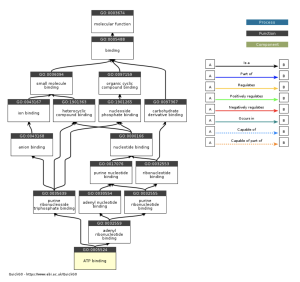
Figure 7: The role that ATP binding plays in desmosomes shows that ATP-related processes may play a role in the development of ARVC. It is possible that it can help develop ARVC through cellular pathways.
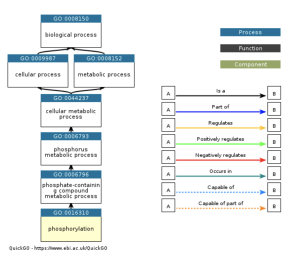
Figure 8: PKP2 can undergo phosphorylation, which may affect the role of signaling and cell adhesion. It can also influence other proteins and the structural integrity of cardiac tissue.
Figure 9: PKP2 does not bind calcium ions directly, but it helps other proteins inside the desmosomes.
DISCUSSION
The main goal of this research was to identify additional genes, pathways, and functions that could contribute to developing a cure for ARVC. This study revealed how it is unclear which genes are up and down regulated (figure 2), and how siNEG and siPKP2 are extremely similar to each other (figure 3). Because these results were vague, it did not support the hypothesis. In order for these results to support the hypothesis, there would need to be more and clearer results. For the DAVID results, while PKP2 is not directly related to calcium signaling pathway (figure 4), chemical carcinogenesis (figure 5), or TNF signaling (figure 6), these pathways can affect its adhesion properties (Novelli, Valeria, et al. 2018). Even though calcium ion binding (figure 9) is indirectly related to PKP2’s function, it can influence the desmosomal function (Chantal J.M., et al 2019). Additionally, PKP2 can undergo phosphorylation (figure 8), which can affect its role in cell adhesion and signaling. Although ATP binding is not directly related to PKP2, it is part of the desmosomes, suggesting that ATP-related processes may depict a role in the development of ARVC (figure 7). It is possible that some cellular processes in PKP2 involve ATP binding proteins. These results align with the hypothesis by providing a better understanding of how gene expression can differ when PKP2 is absent.
Comparing these results to previous studies, the previous studies were clearer because this research was unable to use a clinical setting to find more in depth results. Other studies, like one’s using mouse models, have succumbed to a more in depth understanding of PKP2’s importance (Wu, Iris, et al. 2024). These results from this study can help develop a vaccine or cure for ARVC, especially given the limited data that is currently available in this topic. These results could further assist scientists or other researchers to better understand ARVC.
Limitations
Some limitations are, of course, not having as much information as preferred, and having trouble finding samples to analyze. Since there is reliance on bioinformatics datasets from microarray experiments made by other researchers, one drawback is that the genes that were identified will require additional research in a laboratory or clinical setting before a cure for ARVC can be developed. For future research, the identified genes should be tested in a laboratory to determine if another gene could counteract or prevent the development of ARVC.
Table 1: Summary Table of key gene functions and biological pathways identified in this study
GO results | KEGG results | ARVC connection |
ATP binding | Chemical carcinogenesis – reactive oxygen species | ATP – Mitochondria is the powerhouse of the cell. In ARVC, there may be a dysfunction, leading to less ATP being produced. This results in damage to cardiac cells. ROS = very oxygen reactive molecules. Excessive ROS leads to damaged DNA and proteins. This can also lead to cancer. |
Phosphorylation | Calcium signaling pathway | Abnormal phosphorylation can lead to the development of ARVC. Mutations of desmosomal proteins can lead to arrhythmias, which is when calcium is disrupted |
Calcium ion binding | TNF signaling pathway | In TNF-α, more apoptosis of cardiomyocytes can contribute to ARVC. Disrupted calcium homeostasis can lead to reduced cardiac contraction. |
Conclusion
The goal of this study was to identify gene functions and pathways that can contribute to the development of a cure for ARVC. Regarding the research question, the genes picked were involved in the causes of ARVC and human diseases. The values from GEO2R were analyzed using DAVID, which resulted in GO terms and KEGG pathways. Key GO terms that were acquired included phosphorylation, calcium ion binding, and ATP binding. The KEGG pathways selected were calcium signaling pathway, chemical carcinogenesis – reactive oxygen species, and the TNF signaling pathway. Moreover, regarding the scientific question, how many genes were expressed differently when PKP2 was removed from the heart cells remains unclear. However, the biological pathways identified are closely related to the desmosomes, which helps develop ARVC. The hypothesis was not supported, accentuating that many factors that contribute to ARVC remain understood, likely due to the relatively recent discovery of this disease. Some suggestions for further research are to address other causes of ARVC and then conduct additional studies to develop a treatment for ARVC, since this study’s findings only focused on one aspect of ARVC. Although there is no official cure yet, this study’s findings contribute many insights which may eventually lead to the development of a cure.
References
- “About GEO2R.” (2024). NCBI GEO. https://www.ncbi.nlm.nih.gov/geo/info/geo2r.html
- Cerrone, M., Lin, X., Zhang, M., Agullo-Pascual, E., Pfenniger, A., Chkourko-Guskov, K., …, & Delmar, M. (2017). Plakophilin-2 is required for transcription of genes that control calcium cycling and cardiac rhythm. Nature Communications, 8(1), 1-12. https://www.nature.com/articles/s41467-017-00127-0
- Chantal, J. M., Morais, P., Moreira, S., Leite-Moreira, A. F., & Branco, L. M. (2019). Plakophilin-2 haploinsufficiency causes calcium handling deficits and modulates the cardiac response towards stress. International Journal of Molecular Sciences, 20(17), 4076. https://www.mdpi.com/1422-0067/20/17/4076
- Dalal, D., Nasir, K., Bomma, C., Tandri, H., Dong, J., & Tichnell, C. (2006). Clinical features of arrhythmogenic right ventricular dysplasia/cardiomyopathy associated with mutations in plakophilin-2. Circulation, 113(13), 1641-1649. https://pubmed.ncbi.nlm.nih.gov/16549640/
- Giordano, F. J. (2005). Oxygen, oxidative stress, hypoxia, and heart failure. Journal of Clinical Investigation, 115(3), 500-508.
- He, Y., Rappuoli, R., De Groot, A. S., & Chen, R. T. (2010). Emerging vaccine informatics. Journal of Biomedicine and Biotechnology, 2010, 1-10.
- Joshi-Mukherjee, R., Coombs, W., Izumo, S., & Delmar, M. (2008). Characterization of the molecular phenotype of two arrhythmogenic right ventricular cardiomyopathy (ARVC)-related plakophilin-2 (PKP2) mutations. Heart Rhythm, 5(12), 1715-1723. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2636742/
- Kohan, R., & Pastore, A. (2020). Reactive oxygen species in cancer: A paradox between pro- and anti-tumour activities. Cancer Chemotherapy and Pharmacology, 85(4), 1-10. https://link.springer.com/article/10.1007/s00280-020-04103-2
- Lindsey, S. H., Carbone, J. M., & Lindsey, P. W. (2005). Tumor necrosis factor signaling and the induction of heart failure: What can we learn from mouse models? Current Heart Failure Reports, 2(4), 195-201.
- Maas, W. J., Knook, D. L., & Wisse, E. (2000). Assessment of some critical factors in the freezing technique for the cryopreservation of precision-cut rat liver slices. Cryobiology, 40(3), 261-272. https://pubmed.ncbi.nlm.nih.gov/10860624/
- Novelli, V., Mazzanti, A., Pagan, E., Sibona, M., Robyns, T., & Calore, M. (2018). Pleiotropic phenotypes associated with PKP2 variants. Frontiers in Cardiovascular Medicine, 5, 184. https://www.frontiersin.org/articles/10.3389/fcvm.2018.00184/full
- Pérez-Hernández, M., Tapia, M., Mechulan, A., & Álvarez, D. (2021). Transcriptomic coupling of PKP2 with inflammatory and immune pathways endogenous to adult cardiac myocytes. Frontiers in Physiology, 11, 623190. https://www.frontiersin.org/articles/10.3389/fphys.2020.623190/full
- Saguner, A. M., Brunckhorst, C., & Duru, F. (2014). Arrhythmogenic ventricular cardiomyopathy: A paradigm shift from right to biventricular disease. World Journal of Cardiology, 6(4), 154-174.
- Stanford Health Care. (2017). Symptoms of arrhythmogenic right ventricular dysplasia/cardiomyopathy. https://stanfordhealthcare.org/medical-conditions/blood-heart-circulation/arrhythmogenic-right-ventricular-dysplasia-cardiomyopathy/symptoms.html
- Wu, I., Loyer, X., Simard, F., Bortnick, A., & Seidman, C. (2024). AAV9
improves heart function and survival in a PKP2-deficient mouse model of arrhythmogenic right ventricular cardiomyopathy. Nature Communications, 9, 450. https://www.nature.com/articles/s43856-024-00450-w - Wu, I., Mereles, D., Meder, B., & Giannitsis, E. (2024). GEO accession viewer: GSE253223. NCBI GEO. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE253223
- Zorio, E., Medina, P., Rueda, J., Millán, J. M., de la Calle, G., & Gilabert-Estellés, J. (2009). Insights into the molecular basis of familial cardiomyopathies: From bench to bedside. Applied Microbiology and Biotechnology, 85(1), 1-18.



