Using Bioinformatics to Uncover the Gene Expression of Genes in Affected and non Affected Patients with Cystic Fibrosis
ABSTRACT
Background
Cystic fibrosis (CF) is a genetic disorder caused by mutations in the CFTR gene, leading to the production of thick, sticky mucus that primarily affects the lungs and digestive system. This results in chronic respiratory infections, difficulty breathing, digestive issues, and reduced life expectancy. While treatments like CFTR modulators have improved outcomes, challenges remain, including limited access to therapies, progressive lung damage, and the lack of treatments for rare mutations. Researchers aim to better understand CFTR gene expression to develop targeted therapies, explore gene-editing technologies, and personalize treatments, ultimately improving quality of life and reducing the burden of this disease.
Methods
Bioinformatics tools and databases were used to analyze MRSA data. Data was collected from the National Center for Biotechnology Information. Gene Expression Omnibus (GEO), which included 2 groups, affected and non affected. Differentially expressed genes (DEGs) were identified by removing data with a p-value higher than 2.20e-12. The top 50 DEGs were analyzed using ShinyGO, which incorporated KEGG.
Results
The original total number of genes was 18331, which was then analyzed down to the top 50 DEG’s, with major expression across the group. The use of ShinyGO showed 2 key pathways: Cell adhesion Molecules, and Asthma which showed key genes such as MHC, NLGN4Y, MHC-2, which are associated with inflammation and immune responses.
Conclusion
The study finds genes that could be markers of cystic fibrosis in patients and indicate inflammation and immune deficiencies, These genes can be later tested in laboratories to find possible therapies for CF
KEYWORDS: Cystic fibrosis, MRSA, NCBI, GEO, KEGG, GO, MHC, NLGN4Y, MHC-2, Cell adhesion molecules, Asthma
INTRODUCTION
Cystic Fibrosis is an autosomal recessive disorder that targets the CFTR protein causing the build up of Air Surface Liquid (ASL). These fluids, also called secretions, are usually thin and slippery to protect the body’s internal tubes and ducts and make them smooth pathways, But in people with CF, a changed gene causes the secretions to become sticky and thick. This causes the blocking of pathways to locations such as the lungs, digestive tract, and other organs (1)
What can be done to curve the significant abnormalities when it comes to CFTR. Understanding the Gene in the long arm of chromosome 7. (1) The gene mutation that causes CF has been discovered but currently researchers must identify ways of affecting said gene. Since 2012, recent approaches have enabled the identification of small molecules targeting either the CFTR protein directly or its key processing steps, giving rise to novel promising therapeutic tools yet there is nothing concrete.(2)
The disease is genetic in nature and is most commonly found in the caucasian race more than any other race, and remains one of the most common life-shortening disorders in the white population, with an estimated median survival age of 33.4 years in the US. There is no cure but options may become more available due to continued study of the disorder. (2)
It occurs due to a mutation in the long arm of Chromosome 7 and focuses on cells that produce mucus and sweat in the body. This causes issues with a protein called CFTR.(1)
Patients with the same CFTR mutation can exhibit vastly different disease severities. This variability may be influenced by modifier genes, epigenetics, and environmental factors, which are not fully understood.
The goal of this research is to use bioinformatics to identify and expand the knowledge of cystic fibrosis (CF). Specifically, we used NCBI GEO, ShinyGo, KEGG, and GO bioinformatics tools.NCBI GEO2R is an interactive online tool provided by the National Center for Biotechnology Information that allows users to compare two or more groups of samples in a Gene Expression Omnibus (GEO) dataset to identify differentially expressed genes. In addition, ShinyGo, KEGG, and GO bioinformatics tools were used to further analyze the results obtained from GEO2R. ShinyGo is an online platform for statistical and graphical visualization of biological data, helping to explore patterns, correlations, and trends in high-throughput data.
KEGG (Kyoto Encyclopedia of Genes and Genomes) is a comprehensive database that integrates genomic, chemical, and systemic functional information, enabling the understanding of high-level functions and utilities of biological systems, such as pathways and networks (14).GO (Gene Ontology) is a framework for the standardized representation of gene and gene product attributes across species and databases, providing a structured vocabulary for biological processes, cellular components, and molecular functions (15).These bioinformatics tools together allowed for a comprehensive analysis and interpretation of the datasets, contributing to the expansion of our understanding of CF-related genetic and molecular mechanisms.
Our hypothesis is that there will be a significant difference in gene expression when we compare healthy and infected samples. Advancements in CF treatment have significantly increased life expectancy, which was once only a few years post-diagnosis. Continued research can help further extend and improve the quality of life for patients.
METHODS
Collection of Datasets
GEO2R (www.ncbi.nlm.nih.gov/geo/geo2r/) is an online tool designed for comparing and analyzing gene expression across different sample groups. It shows data as visual data charts Accessed the NCBI library. I used the GEO database to search for cystic fibrosis microarray data. I identified a dataset from research on cystic fibrosis that found that CFTR deficiency in cystic fibrosis disrupts lipid metabolism, promotes chronic oxidative stress, and drives a persistent pro-inflammatory state in airway epithelial cells. I then processed this GEO dataset for my study.

Figure 1: Methods Flow chart. This flow chart shows the methodological approach used in this research.
Data Analysis of GEO2R Results
To process the datasets, I first defined or categorized the data into two groups; affected patients and non-affected patients, and then utilized the no-code GEO2R ‘Analyze’ tool, which operates with pre-programmed R scripts. Analysis with this tool generated multiple graphs of gene expression results which we analyzed. Using the volcano plot, I examined gene expression differences between the various sample groups. From the Venn diagram results, I evaluated the number of expressed genes and identified overlapping genes among the different human sample groups.
Narrowing down the number of differentially expressed genes (DEGs) to top 50
Next, I applied statistical analysis to identify the top 50 most significantly expressed genes from my samples. I downloaded the top differentially expressed genes (DEGs) data into a Google Sheet and selected the top 50 DEGs by removing any data with a p-value higher than 2.20e-12.
Further Analysis of Top Differentially Expressed Genes using ShinyGO, KEGG and GO Bioinformatics tools
To analyze the top 50 identified DEGs and determine the functions and participating key pathways for key genes, I used the ShinyGo bioinformatics tool. ShinyGO is a online bioinformatics program that allows for the creation of KEGG pathway visualizations when imputing large sets of gene data. ShinyGo also generates Gene ontology or GO results to help determine potential functions of key genes from the top 50 DEGs. Altogether, ShinyGo, the GO bioinformatics and KEGG databases (7, 14, 15) were used to find out what the top genes potentially do in patients affected and non-affected with Cystic Fibrosis
RESULTS
Identification of Genes Expressed Differently in the CF Samples
The GEO2R bioinformatics tool was utilized to identify differentially expressed genes (DEGs). The results were visualized using a volcano plot and a Venn diagram shown in Figure 2. In the volcano plot comparing affected versus non affected versus. The genes were observed to be differentially expressed. A portion of the genes were very even, while a slight number displayed lower expression levels. In the plot, red dots represent highly expressed (upregulated) genes, blue dots indicate less expressed (downregulated) genes. The venn Diagram produced a total of 7229 genes expressed between each other out of the total of 18331 genes in the data set.

Figure 2 Volcano diagram and Venn diagram. (A) The volcano map of GSE154802 . (B) The volcano map of GSE40611. Upregulated genes are colored in red; downregulated genes are colored in blue. (C) The two datasets showed an overlap of 7229 genes.
Statistical Analysis of (50) Differentially Expressed Genes (DEGs)
To identify the top 50 DEG’S from 18331 total any p-value greater than 2.20e-12 was excluded. geo2r data.
Enrichment Analysis to Determine Potential Functions of the Identified Genes
Through ShinyGO I identified two pathways, Cell adhesion Molecules and asthma from the KEGG analysis. Through this I identified the key genes of MHC-2, and NLGN4Y (Figure 3)
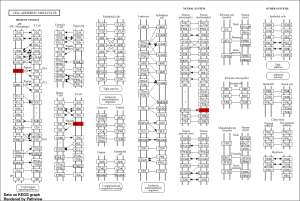
FIGURE 3: Key pathway and key genes associated with the top 50 DEGs: Cell adhesion Molecules. Key genes highlighted in Red: MHC-2, and NLGN4Y
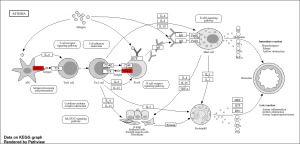
FIGURE 4: Key pathway and key genes associated with the top 50 DEGs: Asthma. Key genes highlighted in Red: MHC II
DISCUSSION
The main goal of the research paper is to identify genes expressed differently in individuals with cystic fibrosis and without. During which genes related to cell adhesion and Asthma (Figure 3 and 4) both exhibited severe differences compared to each other. The original hypothesis of the study was that there would be significant differences in expression of genes which was half proven correct due to the findings showing few gene differences but rather major differences (Figure 2).
The findings of our the study (Table 1) are backed up by other studies as several discuss CF’s relation to the epithelial cells (4). First, using ShinyGo functional and enrichment analysis bioinformatics tool, our study identified cell adhesion molecules and Asthma as the key pathways associated with the top 50 DEGs identified.
Table 1: Summary of the prevalent genes and enriched pathways found in this analysis analysis
Key Genes | Key Pathways | Connection to Cystic Fibrosis |
MHC-II | Cell adhesion Molecules/Asthma | MHC-II molecules are critical for presenting antigens to CD4+ T cells, facilitating the adaptive immune response. In CF, chronic lung infections (e.g., with Pseudomonas aeruginosa or Staphylococcus aureus) lead to heightened immune activation. Variations in MHC-II genes can influence immune response intensity and may determine susceptibility to recurrent infections and inflammation in CF patients. |
NLGN4Y | Cell adhesion Molecules | Neuroligins are synaptic adhesion molecules involved in neural development and function. While NLGN4Y is not directly implicated in the pathology of CF, its relevance may emerge in studying CF-associated comorbidities, such as cognitive or neurodevelopmental issues linked to chronic hypoxia or inflammation. For instance, chronic illness and systemic inflammation in CF could indirectly affect neurological development or function, where molecules like NLGN4Y might play a modulatory role. |
MHC | Asthma | MHC system or an immune-related protein, its role could involve modulating the immune response in CF |
Cystic fibrosis has been documented to be linked to cell adhesion molecules (11). Adhesion molecules are cell surface proteins that are involved in binding cells to one another with an extracellular matrix (ECM) (16). In layman’s terms it acts like molecular glue helping cells stick to one another. For example, ATS journal noted in a study how the adhesion molecules may be linked to an increase in inflammation in CF patients (11). The NLGN4Y gene is defined as a key regulator of cell adhesion and immune response modulation, playing a critical role in cellular interactions within the extracellular environment (12). It functions by encoding a protein that facilitates intracellular binding and immune signaling. The connection of NLGN4Y to CF is that its altered expression may contribute to heightened inflammation and chronic infections seen in CF patients, particularly in the lungs, by affecting the adhesion properties of epithelial cells and immune cell interactions (12).
The MHC-2 class molecule facilitates the adaptive immune response by presenting antigens to T-cells. (Its major connection to CF is that, when patients with CF experience chronic lung infections (e.g., with Pseudomonas aeruginosa or Staphylococcus aureus), it leads to heightened immune activation. This persistent immune response can contribute to lung damage and disease progression. Further research in laboratory settings is needed to fully elucidate this connection. Asthma was another key pathway identified in our study. Asthma is defined as a chronic inflammatory airway disease characterized by reversible airway obstruction, bronchial hyperresponsiveness, and increased mucus production (13). It connects to CF because both conditions involve airway inflammation, mucus hypersecretion, and immune dysregulation. This also requires further testing in a laboratory environment.
Conclusion and Future Direction
By studying these key pathways and genes, researchers can uncover potential target pathways for personalized interventions, such as immunomodulators or anti-inflammatory therapies, to reduce complications in CF. The identified genes can be tested in the laboratory by scientists in the laboratory or clinical trials to determine if possible cures can be identified from it.
Limitations
In our case, since we rely on bioinformatics datasets derived from microarray experiments conducted by other researchers, a key limitation is that the identified genes will require further investigation in laboratory or clinical settings before progressing toward vaccine development.
References
- Gibson, R. L., Burns, J. L., & Ramsey, B. W. (2003). Pathophysiology and management of pulmonary infections in cystic fibrosis. American journal of respiratory and critical care medicine, 168(8), 918-951.
- Fanen, P., Wohlhuter-Haddad, A., & Hinzpeter, A. (2014). Genetics of cystic fibrosis: CFTR mutation classifications toward genotype-based CF therapies. The international journal of biochemistry & cell biology, 52, 94-102.
- Castellani, C., & Assael, B. M. (2017). Cystic fibrosis: a clinical view. Cellular and molecular life sciences, 74(1), 129-140.
- Rommens, J. M., Iannuzzi, M. C., Kerem, B. S., Drumm, M. L., Melmer, G., Dean, M., … & Collins, F. S. (1989). Identification of the cystic fibrosis gene: chromosome walking and jumping. Science, 245(4922), 1059-1065.
- Saiman, L., & Siegel, J. (2004). Infection control in cystic fibrosis. Clinical microbiology reviews, 17(1), 57-71.
- Tanya Barrett, Stephen E. Wilhite, Pierre Ledoux, Carlos Evangelista, Irene F. Kim, Maxim Tomashevsky, Kimberly A. Marshall, Katherine H. Phillippy, Patti M. Sherman, Michelle Holko, Andrey Yefanov, Hyeseung Lee, Naigong Zhang, Cynthia L. Robertson, Nadezhda Serova, Sean Davis, Alexandra Soboleva, NCBI GEO: archive for functional genomics data sets—update, Nucleic Acids Research, Volume 41, Issue D1, 1 January 2013, Pages D991–D995, https://doi.org/10.1093/nar/gks1193
- Minoru Kanehisa, Susumu Goto, Shuichi Kawashima, Akihiro Nakaya, The KEGG databases at GenomeNet, Nucleic Acids Research, Volume 30, Issue 1, 1 January 2002, Pages 42–46, https://doi.org/10.1093/nar/30.1.42
- Tang D, Chen M, Huang X, Zhang G, Zeng L, Zhang G, et al. SRplot: A free online platform for data visualization and graphing. PLoS ONE [Internet]. 2023 Nov 9 [cited 2025 Jan 13];18(11):e0294236–6. Available from: https://pubmed.ncbi.nlm.nih.gov/37943830/
- Smith B, Williams J, Schulze-Kremer S. The ontology of the gene ontology. AMIA Annu Symp Proc. 2003;2003:609-13. PMID: 14728245; PMCID: PMC1480173.
- C;, Marion CR;Izquierdo M;Hanes HC;Barrios. “Asthma in Cystic Fibrosis: Definitions and Implications of This Overlap Syndrome.” Current allergy and asthma reports. Accessed January 21, 2025. https://pubmed.ncbi.nlm.nih.gov/33560464/.
- Circulating adhesion molecules in cystic fibrosis | American Journal … Accessed January 21, 2025. https://www.atsjournals.org/doi/full/10.1164/ajrccm.157.4.9704134.
- “NLGN4Y Neuroligin 4 Y-Linked [Homo Sapiens (Human)] – Gene – NCBI.” National Center for Biotechnology Information. Accessed January 21, 2025. https://www.ncbi.nlm.nih.gov/gene/22829.
- Jesenak M;Durdik P;Oppova D;Franova S;Diamant Z;Golebski K;Banovcin P;Vojtkova J;Novakova E; “Dysfunctional Mucociliary Clearance in Asthma and Airway Remodeling – New Insights into an Old Topic.” Respiratory medicine. Accessed January 21, 2025. https://pubmed.ncbi.nlm.nih.gov/37516275/.
- Kanehisa M;Furumichi M;Sato Y;Kawashima M;Ishiguro-Watanabe M; “Kegg for Taxonomy-Based Analysis of Pathways and Genomes.” Nucleic acids research. Accessed January 26, 2025. https://pubmed.ncbi.nlm.nih.gov/36300620/.
- Ashburner M;Ball CA;Blake JA;Botstein D;Butler H;Cherry JM;Davis AP;Dolinski K;Dwight SS;Eppig JT;Harris MA;Hill DP;Issel-Tarver L;Kasarskis A;Lewis S;Matese JC;Richardson JE;Ringwald M;Rubin GM;Sherlock G; “Gene Ontology: Tool for the Unification of Biology. the Gene Ontology Consortium.” Nature genetics. Accessed January 26, 2025. https://pubmed.ncbi.nlm.nih.gov/10802651/.
- Alberts, Bruce. “Cell-Cell Adhesion.” Molecular Biology of the Cell. 4th edition., January 1, 1970. https://www.ncbi.nlm.nih.gov/books/NBK26937/.



