Using Bioinformatics to Identify Pathways for Deinococcus Radiodurans while under Environmental Stress
ABSTRACT
Background
Deinococcus radiodurans is an extremophilic bacterium known for its remarkable resistance to radiation and DNA damage. It also plays a promising role in heavy metal remediation, particularly in cleaning radioactive waste. However, the genetic mechanisms behind its stress resistance, especially under cadmium chloride (CdCl₂) exposure—are not fully understood. Understanding these pathways could enhance its use in environmental and medical applications.
Methods
With the GEO dataset GSE20383 containing 16 CdCl₂-treated and 16 control D. radiodurans, DEGs were identified with GEO2R. The top 30 DEGs were sorted according to p-value (≤ 0.05) and log fold change. GO enrichment was derived with ShinyGO, which identified functional categories, pathways, along with these genes.
Results
A total of 15 significantly downregulated and 15 significantly upregulated genes were identified among 3,116 DEGs. GO term enrichment showed enriched terms in phosphate transport, glycosaminoglycan binding, and peptidoglycan catabolic processes. These functions suggest potential roles in DNA repair, biofilm formation, and cell wall remodeling—key strategies for D. radiodurans to withstand cadmium chloride stress. The shared enriched pathways point to coordinated stress resistance and structural integrity maintenance, aligning with the study’s goal of uncovering genetic mechanisms that enable the bacterium’s extreme environmental resilience.
ConclusionThis study identified significant pathways responsible for stress resistance against heavy metals in D. radiodurans. Its application could result in its successful use in bioremediation with enhanced efficiency and its successful use in biomedicine against radiation damage with targeting these pathways of expression.
INTRODUCTION
Deinococcus radiodurans is a nonpathogenic bacterium best known for its resistance to DNA damaging causes, such as radiation and UV radiation, as well as its ability to repair DNA effectively and efficiently [1]. Paired with its ability to remediate heavy metals in aqueous wastes [2], D. radiodurans has shown to be a possible solution at removing radioactive waste [2].
The main problem or challenge is understanding the ability of D. radiodurans to rebuild thousands of broken DNA strands [3]. We need to understand what pathways are responsible for its ability to survive under stress, and how further research of these pathways can lead to more effective use in cleaning radioactive waste.
D radiodurans have powerful DNA repair mechanisms like homologous recombination, with pathways such as RecBCD and RecFOR, and extended synthesis-dependent strand annealing. It also produces antioxidant enzymes such as catalases and superoxide dismutases that neutralize oxidative stress [3]. Genomic analysis has suggested that the circular chromosomes of the bacterium are toroidally folded, which may be useful in preserving genetic integrity after desiccation or damage [4]. D. radiodurans can withstand vacuum environments, low temperatures, and starvation regimes—qualities that have made it a model organism in astrobiology and synthetic biology [15]. Current bioinformatics research is continuing to unveil its stress response networks and regulatory mechanisms, which hold the potential to unlock synthetic organism design with built-in stress resistance or the optimization of bioremediation technologies.
To be able to understand how well it can remove heavy metals, we analyzed 16 CdCl₂ treated D.radiodurans and 16 untreated D.radiodurans in the GSE20383 database. Identifying differentially expressed genes (DEGs) through GEO2R, we enriched the top 30 genes with Gene Ontology (GO) to find the top pathways. Through the research of the top pathways in D.radiodurans under heavy metal stress, the function of those pathways are explored and the purpose is theorized.Researchers can use these results to identify the genes correlated to the pathways to further understanding of the organism. This research could lead to further enhancing D.radiodurans in its purpose of clean radioactive waste, but could lead to medications that can withstand radiation.
The goal of the research is to identify genes that are significantly expressed differently in Deinococcus radiodurans while treated with Cadmium Chloride and to determine what functions and biological pathways these genes are enriched in. We hypothesize that there are some genes that are expressed differently and have functions that can point to the ability of the bacteria to withstand high levels of environmental stress.
METHODS
Data Collection and Analysis of GEO2R Data
In this study, the dataset was collected using the National Library of Medicine (NCBI) , the National library for biological articles and databases [9] with the accession code, GSE20383 (Transcriptional profiling of Deinococcus Radiodurans comparing wild type cells with treated cells at four time points).
Data was collected from GEO 2R, a tool used to compare GEO datasets [9]. The datasets were defined into groups of untreated D. Radiodurans and Cadmium Chloride treated D.Radiodurans. These groups were analyzed using GEO2R bioinformatics built in the R programming language, incorporating pre-programmed AI algorithms. Figure 1 provides a detailed summary of the overview of the research steps, the bioinformatics tools and databases employed throughout the study.
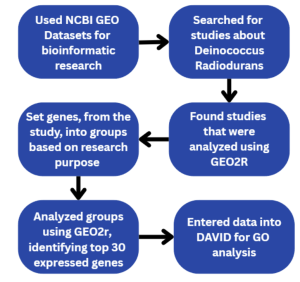
Figure 1: Overview of Research Methodology: A step-by-step outline of the experimental workflow and bioinformatics tools employed in this study, from data collection and analysis of differential gene expression, functional annotation, and pathway enrichment analysis.
Identification of the Top Differentially Expressed Genes
From the GEO 2R results, 3116 genes were identified as differentially expressed. To pinpoint the top 30 most significantly differentially expressed genes in the dataset, we used statistical analysis. This process used p-value, selecting genes that only have less than or equal to 0.05 p-value, to eliminate any expressed genes based on chance; and log fold change, selecting the top 15 and bottom 15, to prioritize the most important genes based on their differential expression across the sample that were compared.
Data Analysis Using SRPlot, KEGG, and GO Bioinformatics Tools
Then Gene Ontology (GO) bioinformatics tools and databases were utilized to analyze the functions of these top genes. Using GO, we analyzed the top 30 expressed genes to uncover pathways and functions that the gene might be involved in.
RESULTS
Identification of Differentially Expressed Genes
The first bioinformatics tool used to identify the top Differentially Expressed Genes (DEGs) was NCBI’s GEO 2R tool. From the first two results very few genes were expressed differently from the treated and the untreated D.radiodurans [Figure 2].
In the volcano plot [Figure 2], the dots represent different genes that were identified and are being compared from both the groups. The red dots show up-regulated genes, genes that were producing more of a specific protein, while the blue dots show down-regulated genes, genes that were producing less of a specific protein. The black dots are genes that were equally expressed in both treated and untreated groups.
In the Venn Diagram [Figure 2], it shows that between the treated and untreated groups, there aren’t any similar genes. Out of the 3116 DEGs, only 7 genes show an overlap between the Cadmium Chloride treated and the untreated D.Radiodurans.
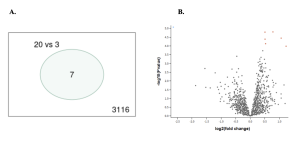
Figure 2: Differentially Expressed Genes: A. The venn diagram, on the left, shows the total number of genes found in this study, as 3116 and 7 genes are common between the two groups. B. The volcano plot, on the right, shows how the genes are being expressed between the CdCl₂ treated and untreated D.Radiodurans groups. The red dots show up-regulation, and the black show equal amount of gene expression or gene regulation between the two groups.
Identification of 30 Statistically Significant Differentially Expressed Genes (DEGs)
The genes collected from the GEO analysis came out to be 3116 genes. To narrow down the genes into only the most expressed genes, we used statistical analysis. Using p-value, we filtered out any genes with a p-value higher than 0.05. This would remove any DEGs that were expressed highly due to chance and not due to the variable, CdCl₂. After filtering out any genes that were based on luck, we used logFC to pick the top 15 and bottom 15 regulated genes, finalizing our new database into 15 up-regulated DEGs and 15 down-regulated DEGs (Top 30 genes).
Potential Functions and Enrichment of the Identified Genes and/or pathways
Submitting the top 30 DEGs into the ShinyGO bioinformatics tool, multiple pathways were identified [Figure 3]. ShinyGo is a tool that performs gene ontology (GO) enrichment analysis and other related analyses for a wide range of organisms[16]. Pathways, Glycosaminoglycan Binding, Phosphate transport, and Peptidoglycan Catabolic Process, had the highest Fold Enrichment [Figure 3a].
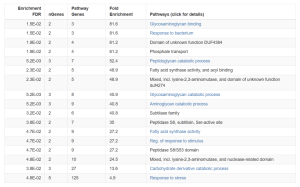
Figure 3a. The table shows enriched biological processes (BP) and molecular functions (MF) identified from the top 30 expressed genes from (site number/experiment name) set using ShinyGO. Columns include the adjusted p-value, number of genes from the input list matching each pathway, total genes in each pathway, and fold enrichment. Pathways such as Glycosaminoglycan binding, Peptidoglycan catabolic process, and Response to bacterium show high enrichment, suggesting functional relevance in the gene list.
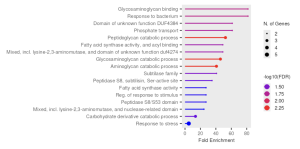
Figure 3b. The plot visualizes the GO enrichment analysis of the input gene set. Each point represents an enriched GO term, with the x-axis showing fold enrichment. Dot size corresponds to the number of genes in the user’s list contributing to the term, and color indicates statistical significance.

Figure 4. Shows the enriched pathways that have similar genes. The closer they are the more genes they share. The Glycosanimoglycan binding is closely related to the Peptiodoglycan catabolic progress, while the phosphate transport is the opposite and not closely related in terms of similar genes. The larger the dots are, the more significant the p-value.
DISCUSSION
Summary of Findings
The goal of the research was to find the most expressed genes while D.radiodurans is submerged in Cadmium Chloride, a heavy metal that D.radiodurans is known for reducing. In the study, we found that Phosphate transport,Glycosaminoglycan Binding, and Peptidoglycan Catabolic Process were found to be the most enriched gene ontology terms [Figures 3b] .
Interpretation of Results
From the GO results, the Glycosaminoglycan Binding, Phosphate transport, and Peptidoglycan Catabolic Process were highly enriched, when under the stress of heavy metals [Figures 3]. Glycosaminoglycans (GAGs) are long unbranched polysaccharides consisting of repeating disaccharides[11]. Known for its viscous properties, they are key components in the ECM providing structural support and facilitating movement of nutrients that could aid in the cell wall strength preventing the CdCl₂ from breaking it down[10].
GAG binding, the interaction between GAGs and proteins, is more complex and has many vital purposes including homeostasis[11]. GAGs are primarily located within the ECG, where it aids in the cell’s structure, however; GAG binding proteins can aid in the production of EPS(extracellular polymeric substances), a thick biofilm, by binding to environmental GAGS [17]. This would create a barrier protecting itself from external stress, CdCl₂ in this case.
In terms of phosphate transport enriched among our top 30 genes [Figures 3], Phosphorus can react with Cadmium to form a precipitate [9], forming a barrier from the CdCl₂. While possible, it is unsure whether CdCl₂ will cause the same reaction with Phosphate. The possibility of the biofilm due to the reaction of phosphorus and Cadmium Chloride can create a precipitate acting as a biofilm to protect itself from external stress.
Lastly, Peptidoglycan Catabolic Process pathways, the breakdown and recycling of peptidoglycan molecules[18], could act as another line of defense to protect itself from external stress. While CdCl₂ could weaken the cell wall, the high enrichment of these pathways [Figure 3], could lead to the possibility of it recycling the broken cell walls and rebuilding it.
Table 1: Summary Table of Key Pathways Identified and their Connection to the Mechanisms in D. radiodurans
Pathway | Function and their connection to D.Radiodurans ability to survive environmental stress |
Glycosaminoglycan binding | Possible usage for the formation of Biofilms when under oxidative stress. |
Phosphate Transport | Possible usage for the repair of DNA strands, or the formation of a precipitate that acts as a barrier between the cell and the environment |
Peptidoglycan catabolic process | Help rebuild the cell wall by recycling broken parts of the wall. |
Comparison with Previous Studies
In other bioinformatics studies, it was found that D.radiodurans create biofilms when under harsh environmental stress. They found that genes DrRRA and drBON1 are crucial to its biofilm regulation[6]. In another study, they found the use of polyphosphate along with the help of manganese ions to protect itself when under oxidative stress, using Hydrogen Peroxide [7]. Similar to the peroxide in the study, CdCl₂ had been found to cause Oxidative Stress in cells[8] aiding in the possibility of phosphate use for D.radiodurans protection. Enzymes such as PPK are upregulated converting ATP into polyphosphate along with Mn+ to prevent proteins from denaturing[7].
Implications
Given the many applications D.radiodurans is being studied for, further knowledge about how it survives various environmental stresses can further increase the possibility of seeing D.Radiodurans in waste cleaning centers while decreasing the time it takes to get there. Furthermore, D.Radiodurans can be used for medical purposes in medications or vaccines[9] that need to undergo high radiative levels such as cancer patients.
Limitations
One of the biggest limitations while doing research about D.radiodurans, is that there isn’t enough research done to find KEGG pathways with the certain genes provided. This prevented any definitive gene that could be pinpointed and researched further upon. Additionally, the bioinformatics datasets from microarray experiments were conducted by other researchers, thus further primary research on phosphate transport and GAG binding will be needed before seeing any real-world application.
Future Directions
The identified pathways and the top 3pathways can be tested in the laboratory for further understanding of its cellular use and how to improve current efforts to use D.radiodurans in real-world situations.
References
- Slade, D., & Radman, M. Oxidative stress resistance in Deinococcus radiodurans. Microbiology and Molecular Biology Reviews, 2011 https://journals.asm.org/doi/full/10.1128/mmbr.00015-10
- Misra CS, Appukuttan D, Kantamreddi VSS, Rao AS, Apte SK. Recombinant D. radiodurans cells for bioremediation of heavy metals from acidic/neutral aqueous wastes. Bioeng Bugs. 2012 Jan;3(1):44–48. doi:10.4161/bbug.3.1.18878. https://www.tandfonline.com/doi/full/10.4161/bbug.3.1.18878
- The radioresistant and survival mechanisms of Deinococcus radiodurans. Radiation Medicine and Protection. 2023 Mar;3(??):?? (review article). doi:10.1016/j.radmp.2023.03.001. https://www.sciencedirect.com/science/article/pii/S2666555723000175
- Levin‑Zaidman S, Englander J, Shimoni E, Sharma AK, Minton KW, Minsky A. Ringlike structure of the Deinococcus radiodurans genome: a key to radioresistance? Science. 2003 Jan 10;299(5604):254–256. doi:10.1126/science.1077865. https://pubmed.ncbi.nlm.nih.gov/12522252/
- Guo Q, Zhan Y, Zhang W, Wang J, Yan Y, Wang W, Lin M. Development and Regulation of the Extreme Biofilm Formation of Deinococcus radiodurans R1 under Extreme Environmental Conditions. Int J Mol Sci. 2024 Jan 1;25(1):421. doi:10.3390/ijms25010421. https://www.mdpi.com/1422-0067/25/1/421
- Dai S, Xie Z, Wang B, Yu N, Zhao J, Zhou Y, Hua Y, Tian B. Dynamic polyphosphate metabolism coordinating with manganese ions defends against oxidative stress in the extreme bacterium Deinococcus radiodurans. Appl Environ Microbiol. 2021 Jul;87(7):e02785‑20. doi:10.1128/AEM.02785‑20. https://journals.asm.org/doi/full/10.1128/aem.02785‑20
- Man Y, Liu Y, Xiong C, Zhang Y, Zhang L. Non‑Lethal Concentrations of CdCl₂ Cause Marked Alternations in Cellular Stress Responses within Exposed Sertoli Cell Line. Toxics. 2023 Feb 9;11(2):167. doi:10.3390/toxics11020167. PMC9962571. https://pubmed.ncbi.nlm.nih.gov/36851042/
- Zhang HY, Li XJ, Gao N, Chen LL. Antioxidants used by Deinococcus radiodurans and implications for antioxidant drug discovery. Nat Rev Microbiol. 2009 Jun;7(6):476 (Reply to Daly’s correspondence ×476). doi:10.1038/nrmicro2073‑c1. https://www.nature.com/articles/nrmicro2073‑c1
- Gene Expression Omnibus (GEO) Series GSE29516. Gene Expression Omnibus Database. 2011 Nov 1; Accession GSE29516. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE29516. Data deposited in NCBI GEO.The data analyzed in this study were obtained from the NCBI Gene Expression Omnibus (GEO) database (GSE29516)
- Jin, M., Xiao, A., Zhu, L., Zhang, Z., Huang, H., & Jiang, L. The diversity and commonalities of the radiation‑resistance mechanisms of Deinococcus and its up‑to‑date applications. AMB Express, 2019. https://pmc.ncbi.nlm.nih.gov/articles/PMC6722170/
- Dai, S., Chen, Q., Jiang, M., Wang, B., Xie, Z., Yu, N., Zhou, Y., Li, S., Wang, L., Hua, Y., & Tian, B. Colonized extremophile Deinococcus radiodurans alleviates toxicity of cadmium and lead by suppressing heavy metal accumulation and improving antioxidant system in rice. Environmental Pollution, 2021. https://www.sciencedirect.com/science/article/abs/pii/S0269749121007090?via%3Dihub
- Authors unknown. Glycosaminoglycan (GAG)-Polysulfate. In: ScienceDirect Topics – Medicine and Dentistry. [Title of section or entry, if specified]. Accessed August 8, 2025. https://www.sciencedirect.com/topics/medicine-and-dentistry/glycosaminoglycan-polysulfate.
- Easy Biology Class. Glycosaminoglycans – Structure, Examples and Functions – Biochemistry Lecture Notes. https://easybiologyclass.com/glycosaminoglycans-structure-examples-and-functions-biochemistry-lecture-notes/. Accessed July 31, 2025.
- Zhou X, Zhang X, Wu X, Zhang Y, Yu X. Response of Deinococcus radiodurans transcriptome to gamma radiation stress. Res Microbiol. 2020 Jan-Feb;171(1):45–54. https://pubmed.ncbi.nlm.nih.gov/31719231/
- Ott E, Kawaguchi Y, Özgen N, Yamagishi A, Rabbow E, Rettberg P, Weckwerth W, Milojevic T. Proteomic and Metabolomic Profiling of Deinococcus radiodurans Recovering After Exposure to Simulated Low Earth Orbit Vacuum Conditions. Front Microbiol. 2019 Apr 29;10:909. doi:10.3389/fmicb.2019.00909. PMCID: PMC6501615. https://www.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2019.00909/full
- Ge, S.X., Jung, D. & Yao, R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics 36, 2628–2629 (2020).
- Guo Q, Zhan Y, Zhang W, Wang J, Yan Y, Wang W, Lin M. Development and Regulation of the Extreme Biofilm Formation of Deinococcus radiodurans R1 under Extreme Environmental Conditions. Int J Mol Sci. 2024 Jan 1;25(1):421. doi:10.3390/ijms25010421. https://www.mdpi.com/1422-0067/25/1/421 mdpi.com
- El-Araby AM, Fisher JF, Mobashery S. Bacterial peptidoglycan as a living polymer. Science Direct. 2025 Feb;84:102562. https://www.sciencedirect.com/science/article/pii/S1367593124001388.



