Identification of TLR2, C5AR1, and AQP9 as Promising Therapeutic Targets for Sarcopenia through Bioinformatic Analysis
ABSTRACT
Background
Sarcopenia is a progressive disease marked by the loss of muscle mass, strength, and function, leading to poor quality of life in aging populations. Primary causes include systemic inflammation, hormonal changes, and physical inactivity. With a prevalence of up to 50% in individuals over 80, sarcopenia poses a growing public health concern. Despite its impact, limited understanding of its molecular mechanisms hinders the development of effective treatments. This study aims to identify genes and pathways involved in sarcopenia as potential targets for gene therapy.
Method
Data from the NCBI GEO database (GSE226151) was collected and analyzed using GEO2R to identify differentially expressed genes (DEGs) from RNA-seq of skeletal muscle tissue in 60 participants categorized as healthy, pre-sarcopenic, or sarcopenic. Then functional analysis was conducted using Gene Ontology (GO) and KEGG pathway enrichment via SR Plot.
Results
Fifty DEGs were identified (p < 0.00003). Among these, enriched GO biological processes included myeloid leukocyte activation, granulocyte migration, and neutrophil migration. Further, key KEGG pathways included neutrophil extracellular trap (NET) formation, complement and coagulation cascades, malaria, and tuberculosis. Myeloid leukocyte activation and NET formation were most significantly enriched. Finally, this study identified three key upregulated genes, TLR2, C5AR1, and AQP9.
Conclusion
Our findings suggest sarcopenia is linked to a strong pro-inflammatory signature involving innate immune activation; particularly neutrophil recruitment, activation and NET formation. Therapeutic approaches such as gene silencing of TLR2, C5AR1, and AQP9 may reduce inflammation and muscle degradation, providing promising strategies to slow or prevent sarcopenia progression.
INTRODUCTION
Sarcopenia is a disease that can be characterized by the loss of muscle mass and strength with the risk of a poor quality of life (1). It is significant because it is a public health concern with a reported prevalence of 50% in those over 80 and this number is expected to continue to rise (2).
The challenge addressed by the research is the limited understanding of genetic factors contributing to sarcopenia. By understanding these factors, this paper can contribute to the development of possible gene-targeted therapies to prevent or reverse sarcopenia.
The primary causes of sarcopenia include systemic inflammation, hormonal changes (such as decreased testosterone), and physical inactivity, all of which contribute to reduced muscle protein synthesis and impaired muscle regeneration (3)(4)(5)(6). Sarcopenia can increase the risk of falls, fractures, disability, and loss of independence. Sarcopenia also contributes to longer hospital stays, higher healthcare costs, and greater risk of early death (1).
Most current bioinformatics research on sarcopenia has been conducted by comparing it with other diseases, rather than studying it as an isolated condition. For example, research comparing pathogenesis in sarcopenia and osteoarthritis has shown that these conditions share common molecular mechanisms such as the FOXO signaling pathway (7). Another research identified biomarkers and therapeutic targets for individuals with both atherosclerosis and sarcopenia (8).
A major challenge in sarcopenia research is the lack of a standardized biomarker that accurately measures its multiple aspects. Existing tools often lack clear thresholds and show variability across populations, making diagnosis and treatment difficult (9). Adding on, the incomplete understanding of the molecular and cellular processes behind sarcopenia hinders the
identification of effective treatments, highlighting the need for deeper research to develop targeted therapies such as gene therapy (10). Gene therapy refers to a medical approach that treats or prevents disease by modifying a patient’s genes to correct genetic defects (22).
Therefore, the goal of the research is to identify genes and pathways that are involved in the onset of sarcopenia as potential targets for a cure in gene therapy. The scientific question being investigated is “Which genes and pathways are involved in the onset of sarcopenia between healthy, pre-sarcopenic, and sarcopenic individuals to determine potential gene-targeted therapies?”.
Our hypothesis is that there will be a difference in expression of genes and pathways between healthy, pre-sarcopenic, and sarcopenic individuals which can be used for gene-therapy strategies. This research is important because it will help identify genes and pathways linked to sarcopenia, which can lead to better ways to detect and measure the disease. This can make diagnosis clearer and more reliable. It also helps us understand how sarcopenia works, which is needed to develop better treatments like gene therapy to help older adults keep their muscle strength and stay healthy.
METHODS
Data Collection and Analysis of GEO2R Data
In this study, the dataset GSE226151 on sarcopenia was collected from NCBI GEO 2R (11) by using the keywords “sarcopenia microarray”. The Gene Expression Omnibus, or GEO, is a repository maintained by the National Center for Biotechnology Information, or NCBI, and freely distributes microarray, next-generation sequencing (NGS) and other forms of high-throughput functional genomic data. (12). GEO 2R is a web application provided by NCBI which allows users to perform sophisticated R-based analysis of GEO data to help identify and visualize differential gene expression (12).
Then the datasets were defined or categorized into groups of healthy, pre-sarcopenia, and sarcopenia. Healthy age refers to individuals who did not develop sarcopenia (Figure 1). Pre-sarcopenia refers to low muscle mass without impact on muscle strength or physical performance. Sarcopenia refers to individuals with the disease (1). The healthy group has 20 samples, the pre-sarcopenia group has 19 samples, and the sarcopenia group has 19 samples. This was analyzed using the no-code GEO2R bioinformatics tool that uses the R programming language (13). Figure 1 below summarizes the steps and bioinformatics tools used in this study.
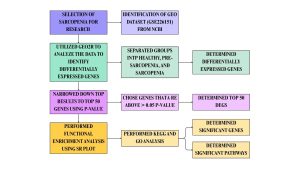
Figure 1: Research Methodology. The flow chart shows a summary of methods and tools used to conduct this research. The research began with data collection and analysis of differentially expressed genes and ended with functional and biological evaluation of the major genes and pathways. The bioinformatics tools and databases used were NCBI GEO for accessing gene expression datasets, GEO2R for identifying differentially expressed genes, and SR Plot for functional enrichment using the GO and KEGG databases.
Identification of the Top 50 Differentially Expressed Genes
Differentially expressed genes refers to genes whose expression levels differ significantly between two or more conditions or groups (14). To identify the most significantly differentially expressed top 50 genes, statistical analysis was applied. This process used p value with a threshold < 0.00003 to prioritize the most important genes based on their differential expression across samples.
Functional and Enrichment Analysis Using SRPlot to Perform GO, and KEGG Analysis
SR Plot was used to perform GO and KEGG analysis to identify the potential functions of the top 50 differentially expressed genes. SR Plot is a web browser that combines data visualization and graphing functions together (15). GO analysis refers to gene ontology enrichment analysis which aids to identify groups of genes that are overrepresented among a list of DEGs. They are grouped into biological processes, cellular components, or molecular formation (16). KEGG analysis refers to the Kyoto Encyclopedia of Genes and Genomes and is used to identify important protein functions, metabolic pathways, signal transduction, and molecular basis of diseases (17). These tools helped uncover the potential roles of pathways and genes in sarcopenia.
RESULTS
Identification of Differentially Expressed Genes
NCBI GEO2R was used to identify differentially expressed genes in my dataset, GSE226151, for sarcopenia. In the volcano plot (Figure 2a and 2b), red dots mean the gene is more expressed and blue dots mean the gene is less expressed. The black/ gray dots means that the gene is not differentially expressed. Results showed that there were no DEGs between pre-sarcopnic and healthy individuals, however, there were numerous genes over and under expressed in groups between pre-sarcopenic vs sarcopenic individuals and sarcopenic vs healthy individuals. This table was used to determine the over-expressed and under-expressed genes.Volcano Plot Signficant Genes
From the venn diagram, the total genes after initial analysis was 16817 (Figure 2c). From the venn diagram, 36 genes were differentially expressed between sarcopenic vs healthy individuals and pre-sarcopenic vs sarcopenic individuals (Figure 2c).
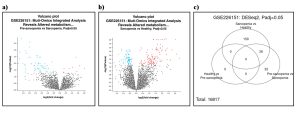
Figure 2: Differentially Expressed Genes. (a) (b)Volcano plots show how the genes are being expressed differently between (a) Pre-sarcopenia and Sarcopenia and (b) Healthy and sarcopenia. The Venn diagram shows the genes that are between pre-sarcopenia, healthy, and sarcopenia patients. Out of 16817 genes that are expressed there is an overlap of genes within the Sarcopenia vs Healthy and Pre-sarcopenia vs Sarcopenia.
Identification of 50 Statistically Significant Differentially Expressed Genes (DEGs)
For the statistically significant genes, a p-value of 0.00003 was used to narrow down the genes. At the conclusion of this analysis, a total of 50 DEGs were identified based on this statistic Top 50 DEGs for Sarcopenia
Potential Functions and Enrichment of the Identified Genes and/or pathways
SR Plot was used to complete functional and enrichment analysis to determine the potential functions of the genes. First, it was determined that the GO group with the highest enrichment score, on average, is biological processes (Figure 3a). After analyzing the Dot Plot for biological process, it was determined that myeloid leukocyte activation had the highest enrichment score as well as p-value (Figure 3b). Then, the genes involved with myeloid leukocyte activation were determined using a c-net plot (Figure 3c).
From the GO results, the genes that stood were the ones involved in myeloid leukocyte activation, these include FGR, LDLR, TLR2, ZC3H12A, CXCR2, PLA2G2A, C5AR1, SLC11A1, and ADGRE2 (Figure 3c).
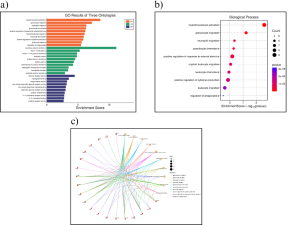
Figure 3: GO Enrichment Analysis (a) The enrichment in all the ontologies from the GO results shows that on average Biological Process (BP) has the most enrichment score. (b) The BP Analysis Dot Plot shows that myeloid leukocyte activation has the most enrichment score and p-value. (c) The BP Analysis C-net pot shows that the genes involved with myeloid leukocyte activation are FGR, LDLR, TLR2, ZC3H12A, CXCR2, PLA2G2A, C5AR1, SLC11A1, and ADGRE2.
From the KEGG results, Neutrophil extracellular trap formation was identified as the most significant pathway because of its high enrichment score and p-value (Figure 4a). Then the C-Net plot and pathway was used to determine significant genes for this specific pathway (Figure 4b and 4c).
From the KEGG results, the important genes that were identified from the C-Net plot for neutrophil extracellular trap formation were CLEC7A, C5AR1, AQP9, FCGR3A, and TLR2 (Figure 4b).The genes that stood out from the neutrophil extracellular trap formation pathway were TLR2, C5aRCLR, FeyRIIIb, and AQP9 (Figure 4c).
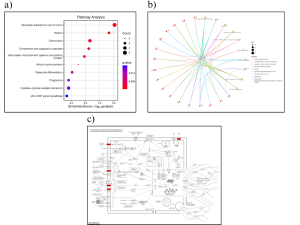
Figure 4: KEGG Enrichment Analysis (a) The KEGG Pathway Analysis Dot Plot shows that neutrophil extracellular trap formation has the most enrichment and p-value. (b) The KEGG Pathway Analysis C-Net Plot shows the various genes involved with neutrophil extracellular trap formation which are CLEC7A, C5AR1, AQP9, FCGR3A, and TLR2. (c) The Neutrophil Extracellular Trap Formation image shows that the significant genes are TLR2, C5aRCLR, FeyRIIIb, and AQP9.
DISCUSSION
Summary of Findings
The main goal of this study was to identify genes and pathways that play a role in the development of sarcopenia to function as potential targets for gene therapy. The pathway identified in this study is neutrophil extracellular trap formation, while the biological process is myeloid leukocyte activation (Figure 3 and 4). By comparing GO and KEGG, TLR2, C5AR1, and AQP9 were identified as significant genes that could contribute to sarcopenia (Figure 3 and 4). After comparing the genes to Log2 fold change values from the volcano plot, it was determined that TLR2, C5AR1, and AQP9 were upregulated (Figure 2).
Table 1: Summary of Key Genes Identified and their Connection to Sarcopenia
Key Genes | Gene Name | GO Process/ KEGG Pathway | Connection to Sarcopenia |
TLR2 | Toll Like Receptor 2 | Myeloid leukocyte activation and neutrophil extracellular trap formation | TLR 2 induces oxidative stress and inflammation which is a cause of sarcopenia (18) |
C5AR1 | Complement C5a Receptor 1 | Myeloid leukocyte activation and neutrophil extracellular trap formation | C5AR1 promotes immune-driven inflammation in muscle tissue which contributes to sarcopenia (19) |
AQP9 | Aquaporin 9 | Neutrophil extracellular trap formation | AQP9 contributes to inflammation which causes sarcopenia (20) |
Interpretation of Results
The pathway that was most significant from our top 50 DEGs in this study, neutrophil extracellular trap (NET) formation, refers to the process by which neutrophils, a type of white blood cell, release NETs. Research has shown that neutrophils must be properly activated to release these NETs. Importantly, the receptor MICL on myeloid cells (myeloid cells are a type of white blood cells and MICL stands for Myeloid Inhibitory C-type Lectin-like Receptor which is a protein receptor) a regulator, preventing excessive NET release. When MICL is blocked or absent, neutrophils become overactive, producing too many NETs, which can lead to inflammation and tissue damage (21). This demonstrates that the biological process of myeloid leukocyte activation plays a critical role in controlling NET formation. Any dysregulation in myeloid activation can result in uncontrolled NET release, contributing to inflammation which is a known factor in sarcopenia. Understanding this connection could inform gene-targeted therapies, specifically by modulating genes including TLR2, C5AR1, and AQP9, which influence inflammatory processes in the musculoskeletal system and may help prevent or treat sarcopenia through gene therapy.
Comparison with Previous Studies
Gene therapy refers to a medical approach that treats or prevents disease by modifying a patient’s genes to correct genetic defects (22). Our study suggests using the genes that impact neutrophil extracellular trap formation to help treat patients with sarcopenia. There have been prior studies that showed the effective use of gene therapy to prevent ag-related sarcopenia. For example, a study demonstrated that AAV1.NT-3 gene therapy effectively prevents age-related sarcopenia in mice (23). Another study found that using rAAV9–SaCas9 to knock out myostatin in aged mice increased muscle mass and fiber size, enhanced satellite cell activity, and modulated TGF-β signaling, highlighting its potential as a gene therapy approach for treating sarcopenia (24). Lastly, a recent study identified ATF3 as a key regulator linking ferroptosis to sarcopenia via the PI3K/Akt signaling pathway, suggesting that targeting ATF3 could prevent muscle cell death and mitigate age-related muscle loss (25).
Implications
The results of this study have significant implications for developing targeted therapies for inflammation in relation to sarcopenia. By identifying TLR2, C5AR1, and AQP9 as overexpressed genes linked to inflammation and NET formation, this research provides candidate targets for gene therapy approaches aimed at reducing chronic inflammation in aging muscle. In practice, therapies could include gene silencing techniques to downregulate these genes, potentially preserving muscle function in older adults (26). Additionally, these findings may inform the development of drugs that block the signaling pathway activated by these genes, offering pharmacological alternatives for patients.
Limitations and Future Directions
In this research, bioinformatics datasets produced from microarray experiments conducted by other researchers were used. As a consequence of this, the identified genes in this study will need to be studied and experimented on in laboratory or clinical environments before it can be used to aid patients through gene therapy. The identified genes can be tested in the lab for laboratory or clinical trials to determine if they can be used for gene therapy to help rehabilitate or cure individuals with sarcopenia.
References
- Cruz-Jentoft AJ, Baeyens JP, Bauer JM, Boirie Y, Cederholm T, Landi F, et al. Sarcopenia: European Consensus on Definition and Diagnosis: Report of the European Working Group on Sarcopenia in Older People. Age and Ageing [Internet]. 2010 Apr 13;39(4):412–23.
- Iannuzzi-Sucich M, Prestwood KM, Kenny AM. Prevalence of Sarcopenia and Predictors of Skeletal Muscle Mass in Healthy, Older Men and Women. The Journals of Gerontology Series A: Biological Sciences and Medical Sciences. 2002 Dec 1;57(12):M772–7.
- Zhao W, Zhang Y, Hou L, Xia X, Ge M, Liu X, et al. The association between systemic inflammatory markers and sarcopenia: Results from the West China Health and Aging Trend Study (WCHAT). Archives of Gerontology and Geriatrics. 2021 Jan;92(33032183):104262.
- Shin MJ, Jeon YK, Kim IJ. Testosterone and Sarcopenia. The World Journal of Men’s Health. 2018;36(3):192.
- Kortebein P, Symons TB, Ferrando A, Paddon-Jones D, Ronsen O, Protas E, et al. Functional Impact of 10 Days of Bed Rest in Healthy Older Adults. The Journals of Gerontology Series A: Biological Sciences and Medical Sciences [Internet]. 2008 Oct 1;63(10):1076–81. Available from: https://academic.oup.com/biomedgerontology/article/63/10/1076/559225
- Cacciatore S, Calvani R, Esposito I, Massaro C, Gava G, Picca A, et al. Emerging Targets and Treatments for Sarcopenia: A Narrative Review. Nutrients [Internet]. 2024 Sep 27;16(19):3271–1. Available from: https://www.mdpi.com/2072-6643/16/19/3271
- Yang J, Zhong J, Du Y, Wang Z, Jiang L, Li Z, et al. Bioinformatics and systems biology approaches to identify potential common pathogenesis for sarcopenia and osteoarthritis. Frontiers in Medicine. 2024 Jun 18;11(38962732).
- Yang J, Zhong J, Du Y, Liu J, Li Z, Liu Y. Bioinformatics analysis based on microarray data reveals molecular crosstalk and immune relationship between sarcopenia and atherosclerosis. Experimental Gerontology. 2025 Jun 14;208:112811–1.
- El-Sebaie M, Walaa Elwakil. Biomarkers of sarcopenia: an unmet need. Egyptian Rheumatology and Rehabilitation. 2023 Sep 11;50(1).
- Najm A, Niculescu AG, Alexandru Mihai Grumezescu, Mircea Beuran. Emerging Therapeutic Strategies in Sarcopenia: An Updated Review on Pathogenesis and Treatment Advances. International journal of molecular sciences. 2024 Apr 12;25(8):4300–0.
- Home – GEO – NCBI [Internet]. Nih.gov. 2019.
- Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, et al. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Research. 2012 Nov 26;41(D1):D991–5.
- https://docs.google.com/document/d/1Ioy2PTjN2C0Dvn-UJCRB1dppcU_WcDm5CiPdccJxKfA/edit?usp=sharing
- Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2009 Oct 24;26(1):136–8.
- Tang D, Chen M, Huang X, Zhang G, Lin Z, Zhang G, et al. SRplot: A free online platform for data visualization and graphing. PLOS ONE. 2023 Nov 9;18(11):e0294236–6.
- Lu Y, Rosenfeld R, Simon I, Nau GJ, Bar-Joseph Z. A probabilistic generative model for GO enrichment analysis. Nucleic Acids Research. 2008 Aug 1;36(17):e109–9.
- Sato N, Miho Uematsu, Fujimoto K, Satoshi Uematsu, Imoto S. ggkegg: analysis and visualization of KEGG data utilizing the grammar of graphics. Bioinformatics. 2023 Oct 1;39(10).
- Kim DS, Cha HN, Jo HJ, Song IH, Baek SH, Dan JM, et al. TLR2 deficiency attenuates skeletal muscle atrophy in mice. Biochemical and Biophysical Research Communications. 2015 Mar 6;459(3):534–40.
- Wang HA, Lee JD, Lee KM, Woodruff TM, Noakes PG. Complement C5a-C5aR1 signalling drives skeletal muscle macrophage recruitment in the hSOD1G93A mouse model of amyotrophic lateral sclerosis. Skeletal Muscle. 2017 Jun 1;7(1).
- Tesse A, Gena P, Rützler M, Calamita G. Ablation of Aquaporin-9 Ameliorates the Systemic Inflammatory Response of LPS-Induced Endotoxic Shock in Mouse. Cells. 2021 Feb 18;10(2):435.
- Malamud M, Whitehead L, McIntosh A, Colella F, Roelofs AJ, Kusakabe T, et al. Recognition and control of neutrophil extracellular trap formation by MICL. Nature [Internet]. 2024 Aug 14 [cited 2024 Aug 15];1–9. Available from: https://www.nature.com/articles/s41586-024-07820-3
- Hanna E, Rémuzat C, Auquier P, Toumi M. Gene therapies development: slow progress and promising prospects. Journal of Market Access & Health Policy [Internet]. 2017 Jan 3;5(1):1265293. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5328344
- Ozes B, Tong L, Myers M, Moss K, Ridgley A, Sahenk Z. AAV1.NT-3 gene therapy prevents age-related sarcopenia. Aging. 2023 Mar 9;
- Weng S, Gao F, Wang J, Li X, Chu B, Wang J, et al. Improvement of muscular atrophy by AAV–SaCas9-mediated myostatin gene editing in aged mice. Cancer Gene Therapy. 2020 May 13;27(12):960–75.
- Chen Z, Hu D, Wu C, Wu Z, Lin J, Liu W. ATF3 as a molecular nexus linking ferroptosis regulation to sarcopenia pathogenesis via PI3K/Akt pathway activation. Experimental Gerontology. 2025 Oct;209:112830.
- El-Sappah AH, Yan K, Huang Q, Islam MdM, Li Q, Wang Y, et al. Comprehensive Mechanism of Gene Silencing and Its Role in Plant Growth and Development. Frontiers in Plant Science. 2021 Sep 7;12.



